Insights on Dual ISH-IHC: Guidance from the experts on performing RNAscope™ ISH and immunohistochemistry on the same sample
- Kristie Wetzel, Bradley Spencer-Dene, Wanda Wang & Mike Millar, Scientist II, Associate Scientist, Research Specialist, Imaging Facility Manager
RNA in situ hybridization (ISH) and immunohistochemistry (IHC) are well-established methods providing unique RNA and protein expression quantification in a morphological context. Often considered complementary technologies, they are bridging the gap between RNA and protein analysis. By targeting different molecules, one the precursor of the other, performing ISH and IHC together can provide complementary information to:
- Identify the origin of secreted proteins – ISH identifies cells producer of the protein, IHC identifies where
- the circulating protein is secreted
- Identify complex tissue structure – in complex structures with multiple cell types, IHC identifies the cell
- type and ISH detects RNA expression inside these cells
- Identify gene expression regulation - translational regulation controls can shut down protein synthesis, or
- protein instability can render IHC ineffective. Analysis of both RNA and protein expression in the same
- tissue allows differentiation between inhibition of transcription and protein instability
- Assess gene therapy – a combination of ISH and IHC can provide a useful comparison between levels
- of transduction and levels of protein. Successful transduction may not always translate to successful
- expression due to regulation or protein instability.
Due to the multiple benefits of running both ISH and IHC on the same sample and same slide with tissues prepared using the same sample preparation method, some of ACD’s RNAscope™ users have begun to develop dual ISH-IHC protocols to perform both analyses on the same slide. There are various ways that these workflow can be set up, as illustrated by Figure 1.
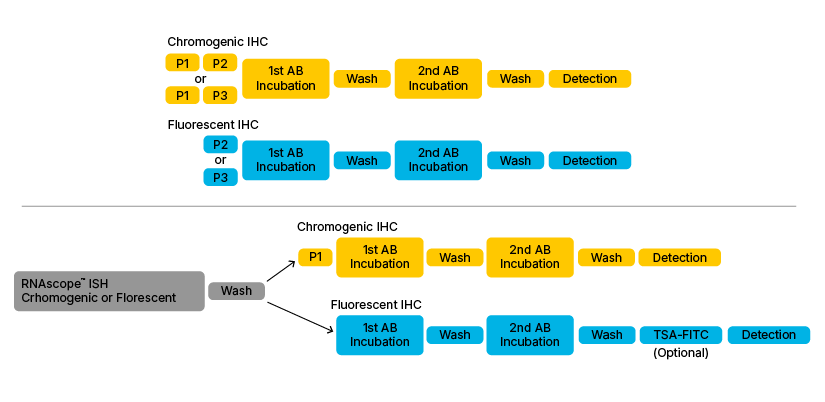
Figure 1: Basic outline of the various Dual ISH-IHC assay combinations. Upper section shows classical IHC workflow when performed individually. Lower section shows the most common dual ISH-IHC workflows found in the literature. P1. P2, P3 and P4 refer to Pre-treatment 1/H2O2 (Hydrogen Peroxide), 2/Target Retrieval Buffer, 3/Protease Plus and 4/Protease IV.
Interview with Kristie Wetzel
Can you tell us about your work and why you developed a dual protocol?
I work as an investigator in the developmental and molecular pathways lab at Novartis. Our group provides immunohistochemistry (IHC) and in situ hybridization (ISH) services not just to our own lab but also to other groups working in diverse disease areas. This means that we get many different sample types depending on the focus of the particular study; it might be tissue from human, mouse, rat... We developed a dual ISH-IHC protocol because fundamentally, we want to be able to see where the mRNA is accumulating. We want to identify which tissue and which cell type is producing it. That’s why RNAscope™ ISH is really important in our work and that’s why we are using it. There are also certain scenarios where our targets don’t have very good antibodies, or using antibodies is difficult such as when profiling a secreted protein. What we get when we combine ISH and IHC is the ability to identify both the particular cell type by IHC and the mRNA profile by ISH. This information is of great interest to us.
How was your dual ISH-IHC protocol set up? Were there any challenges?
We tested a protocol where we first perform the chromogenic RNAscope assay (RNAscope™VS Red) followed by immunofluorescence staining. Our target was a lysozyme protein. Both of the reactions were automated on the DISCOVERY system (Ventana). To set this up, we first confirmed that the RNAscope assay was working on our tissue samples and then we optimized our IHC protocols. We decided to do ISH followed by IHC because it was important to digest the tissue for our lysozyme antibody and the RNAscope protocol has an antigen retrieval step followed by a protease treatment step. It took us a number of weeks and a number of iterations before we really got the protocol optimized but once we did, the resulting staining was really beautiful and provide a interesting data combination to see.
Our major challenge was optimizing the IHC protocol as protein stability is affected by the RNAscope pretreatments. It was important to make sure that the IHC was optimized independently of the RNAscope ISH first before combining it in the dual reaction.
Do you have any recommendations you’d like to share?
I definitely recommend working out the individual protocols separately before combining them. You have to have a good signal individually first, but you also need to realize that even if they’re both working well individually, you’re still going to need to tweak and optimize once you combine them. The IHC protocol is definitely going to need optimizing due to the RNAscope pretreatment - it’s also important to determine the optimal RNAscope pretreatment protocol for each tissue type you have. Once this is established, it’s not going to change. Another thing I would recommend is following the RNAscope protocol to the letter! It works very well if you do this.
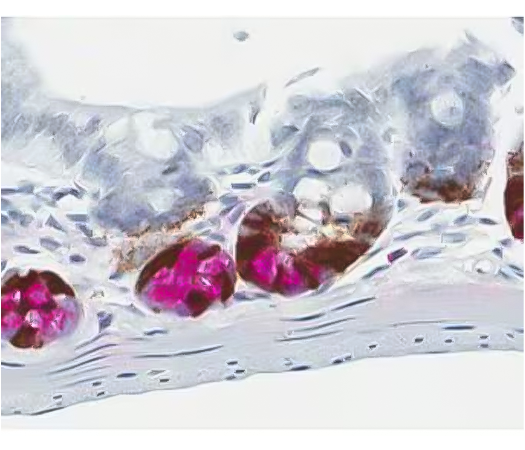
Figure 2. Dual ISH-IHC staining using chromogen labeled enzymes to identify mRNA expression in the stem cells, and protein expression in the Paneth cells in the mouse gut. Provided by Kristie Wetzel.
Interview with Bradley Spencer-Dene
What is the focus of your research?
Originally, I was trained in mammalian embryology, especially my PhD was in craniofacial development, and now I work as a technical specialist within a core pathology service. I have worked extensively on many mouse models for human cancer and disease and my ongoing interest is in the roles of Fibroblast Growth Factors (FGFs) and their receptors in development and disease.
Can you describe your dual ISH-IHC protocol and why you developed it?
Our facility has compiled almost 400 antibodies that work on FFPE mouse tissue and we are regularly being asked to test and optimize new ones. We are also using an increasing amount of antibodies on FFPE human tissue, and even zebrafish now. However, there are many targets that do not have satisfactory antibodies for IHC, so ISH - in particular RNAscope ISH - enabled us to look at the expression patterns and levels of previously unattainable target genes in both mouse and human samples. I was formerly doing both radioactive and non-isotopic ISH for my own studies and as part of my role in the core facility. I started using RNAscope ISH just over a year ago and haven’t looked back.
We decided to develop a dual ISH-IHC protocol due to direct requests from the institute’s scientists, but also in part because of scientific curiosity that drive the innovation.
We are working the protocol manually at the moment but we have the platforms available to take it to automation (DISCOVERY, Ventana). For ISH we are using the chromogenic red kit to take advantage of the natural fluorescence of the Fast Red substrate coupled with Alexa-fluor 488 stained immunofluorescence IHC. We do ISH first followed by IHC. I haven’t tried the reverse on the premise that the mRNA is probably more unstable than the protein epitopes and so would be more likely to degrade following the IHC protocol.
We have tested this on mouse and human normal and tumor tissues plus cell pellets, tissue microarrays and cells grown directly onto glass slides. We have tried five ISH probes (hPPIB, mPPIB, hPD-L1, mLGR6, hZFP36) and 12 antibodies (p-ERK, p-HH3, TTF1, α-1-Na/K-ATPase, F4/80, CAM 5.2 acidic cytokeratins, PD-L1, CD45 common leucocyte common antigen, Cytokeratin 5, Cytokeratin 8, CD3 and active Caspase 3). We chose these as we wanted to look at a variety of cell types, tumor vs lymphocyctic infiltrate as well as proliferative and apoptotic markers. We generally used different targets for ISH and IHC but I did successfully co-stain PD-L1.
Were there any issues and do you have any advice on setting up a protocol?
There are some possible issues with antibodies that normally require trypsin digestion, e.g. F4/80. We found that this antibody didn’t work so you may need to include a trypsin digestion step after ISH at start of IHC protocol. Also, standard methods for reducing/quenching autofluorescence such as 0.1% Sudan Black in 70% EtOH are not compatible with the Fast red RNAscope signal.
I would advise fixing your tissue for 72 hrs at room temperature in 10% Neutral Buffered Formalin and carrying out singleplex RNAscope ISH and IHC staining separately to establish where both signals are and how strong they are alone. We also found it necessary to dilute the RNAscope pretreatment 3 (protease plus) so you might also need to consider optimizing this protease digestion step.
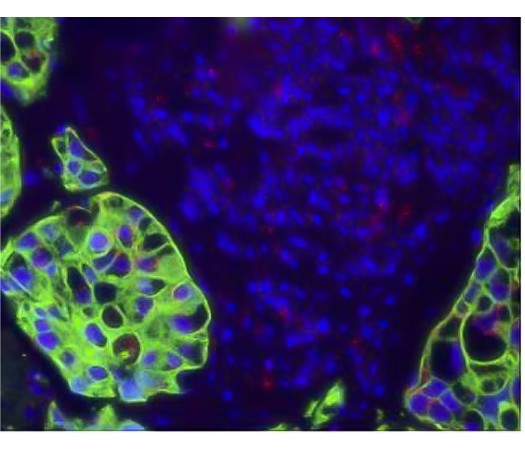
Figure 3. Dual ISH-IHC using fluorescent staining to identify PPI mRNA expression (red dot) and CAM5.2 protein expression (green) in lung sample sample. Provided by Bradley Spencer-Dene.
Interview with Wanda Wang
Please could you tell us about your research and the reason you developed a dual protocol?
Our research group focuses on viral expression in the brain. One of our collaborative projects is a flu model in various animals to investigate flu vaccines. Our collaborators work on the animal infection and we investigate what happens after infection to understand the desease pathology. In another project, we have an animal model for SIV. My particular focus is on in situ data, immunocytochemistry and image analysis, including confocal image analysis and other computer analysis methods.
We are using RNAscope ISH because we want to know where the RNA is located - but we also need to know which cells are infected and which cells are involved. For this reason, we are using IHC for cell identification and RNAscope ISH for detection of cells infected with viral RNA. We are looking for co-localization of these markers, which is important. We developed a dual protocol to be able to get all this data from one slide.
How did you set up your ISH-IHC protocol?
We are looking for viral RNA with the chromogenic red RNAscope kit and cellular markers with immunofluorescence IHC, using Alexafluor 488. We are doing this manually in human and monkey brain FFPE samples. Unfortunately, we don’t have any control over the fixation process of the samples as they originate from other labs, and this can be challenging for pre-treatment conditions. We have set up our protocol to do the ISH first, followed by IHC as the RNA data is the most important to us. I did try to do IHC first followed by ISH but I found that the ISH protocol wiped out my IHC staining but I only tried this once though.
I use the ACD RNAscope protocol as it is, with the provided reagents. Once the in situ is finished, I do immunohistochemistry using the PerkinElmer TSA method. I’ve been able to successfully co-label several different antibodies with ACD RNAscope ISH.
What were the major challenges and do you have any recommendations?
A major challenge for me is protein degradation. I have noticed that I need a higher concentration of antibody in the IHC protocol probably because the protein target is damaged by the ISH protocol.
In terms of recommendations, I’ve found that it’s really important to use pretreatment 3 (Protease plus reagents) provided by ACD. The assay doesn’t work without it. Also, you have to know that your IHC works before you try to combine it in the dual ISH-IHC. For this reason, I find that optimizing the IHC protocol with the RNAscope pretreatments prior to the dual ISH-IHC gives the best results. For the IHC staining, I would recommend using TSA from PerkinElmer to boost the protein signal, and using a high antibody concentration.
Interview with Mike Millar
Please tell us about your work and the SuRF facility.
Our facility provides full core pathology services and microscopic imaging - we’ve been doing this for over a decade now. I’m really a full service scientist in my current position, but prior to that, I was using ISH and IHC for research on somatic genesis. I have an interest in multiplexing using robotic platforms, therefore we have a Leica BOND RX and two BOND MAX machines at the facility.
Why did you develop a dual ISH-IHC protocol and how was this set up?
We developed the protocol for two reasons. One is that we’ve always had an interest in this dual protocol in terms of innovation; and the other is that we want to advance our service and prepare for future needs. Gene expression and immunohistochemistry are naturally complementary and I get the impression from speaking to people that at some point they’re going to want to be able to see dual ISHIHC staining. That’s why we’ve set it up as one of our commercial services.
We’ve developed an automated protocol for Bouin’s fixed testes samples and standard archived FFPE tissues. In our protocol, we do the ISH assay first, followed by the IHC. This was my empirical inclination based on the historic importance placed on RNAse contamination and the general idea that RNA can be easily destroyed. It made sense to me to worry more about the RNA first. We did try to do the IHC first when we were trying out the Ventana Discovery and Leica Bond RX autostainers, but we found that the results were much better when we did the RNAscope assay first.
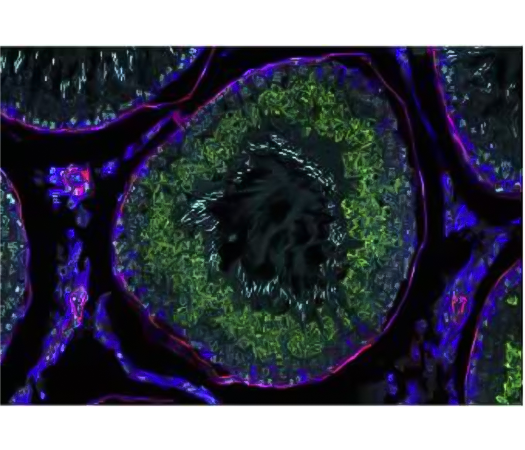
Figure 4 Detection of one mRNA and 2 proteins. Provided by Mike Millar.
One modification we made was to replace the DAB step of the RNAscope protocol with a fluorescein-tyramide, which can be conjugated to FITC or any other color. This way, we can collect fluorescence signals for both the ISH and IHC. We also use a tyramide-based immunofluorescence protocol. We’re getting really great signals using this method as the tyramide detection is probably one of the most sensitive detection methods available.
Are there any challenges with this protocol and do you have any recommendations?
I think the only issue would be if your protein was sensitive to any of the general retrieval steps, which might mean you have to try doing a protein detection first, which isn’t ideal.
I recommend that the first thing you need to do is optimize your RNAscope assay, and then optimize your antibody against these conditions. Sample fixation is crucial and you need to be really consistent with your fixation time. Fixation condition is going to determine the pretreatment when optimizing your RNAscope assay. Then you need to check that the antibody will survive the same pretreatment conditions. Most antibodies are generally okay with stringent antigen retrieval, but there’s probably a handful that aren’t. I would definitely recommend working up both the RNAscope ISH and IHC assays individually first, and then combine them into a standard dual protocol in order to get the best results.
Also, if you’re looking to set-up a dual chromogenic reaction and you think the RNA and protein might co-localize then it would be best not to do DAB first. It has been published that DAB forms an impermeable barrier which would prevent subsequent reagent penetration and target binding. When we do detection using DAB and alkaline phosphatase (ALP) for example, we do ALP first and then DAB. The ALP substrate tends to be more permeable than DAB.
Although our experts perform and set-up their dual ISHIHC protocols differently, they report several common recommendations and advices:
- All Dual ISH-IHC protocols require protocol optimization
- Advisable to perform RNAscope ISH first, followed by IHC
- Advisable to optimize IHC separately using RNAscope pretreatment reagents
- Dual ISH-IHC works better for highly expressed proteins, due to protein degradation during the ISH protocol