Western Blotting of GAPDH in Human Eye Cell Lysate
Human eye cell lysate. Antibody used at 1:500. WB image submitted by a verified customer review.
Western Blot Analysis of GAPDH in Chicken Pectoralis Major
Analysis of GAPDH in chicken pectoralis major using anti-GAPDH antibody. The primary antibody was used at a dilution of 1:4000, incubated for 1 hour at room temperature. Western blot image submitted by a verified customer review.
Western Blot Detection of GAPDH in Multiple Cell Lines and Buffer Protocols
GAPDH-Antibody-1D4-Western-Blot-NB300-221-img0010.jpg
Detection of GAPDH in Western Blot as a Loading Control
GAPDH-Antibody-1D4-Western-Blot-NB300-221-img0011.jpg
Western Blot Detection of GAPDH in Multiple Treated Cell Lines
GAPDH-Antibody-1D4-Western-Blot-NB300-221-img0015.jpg
Western Blotting of GAPDH as a Loading Control for Treated HUVECs
GAPDH-Antibody-1D4-Western-Blot-NB300-221-img0014.jpg
Immunocytochemistry/Immunofluorescence Staining of GAPDH in SH-SY5Y Cells
SH-SY5Y cells stained with GAPDH antibody NB300-221 (green). Nuclear DNA is stained with Hoechst dye (blue).
Simple Western Analysis of GAPDH in Jurkat and MCF-7 Cell Lysates
Lane view shows a specific band for GAPDH using 0.05 mg/mL Jurkat and MCF-7 cell lysates and antibody at 1:25. Electropherogram image of corresponding Simple Western lanes view at WES molecular weight of 40 kDa. Image reported by internal validation.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Knockdown of Ift88 or Kif3a in NP cells does not affect hyperosmotic upregulation of TonEBP. (a) TonEBP/Nfat5 mRNA levels in NP cells with Ift88 knockdown (n ≥ 5). (b) Western blot image showing increased TonEBP expression in response to hyperosmolarity (550 mOsm/kg H2O) independently of Ift88 knockdown. (c) Densitometry analyses of TonEBP with Ift88 knockdown (n ≥ 4). (d) TonEBP/Nfat5 mRNA levels in NP cells with Kif3a knockdown (n ≥ 3). (e) Western blot image showing that hyperosmotic induction of TonEBP is maintained after Kif3a knockdown. (f) Densitometry analyses of TonEBP after Kif3a knockdown (n ≥ 4). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. Western blot images were cropped & acquired under same experimental conditions. See Supplementary Fig. S1-1 for un-cropped Western blot images. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31664118), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - OPN induces EndoMT. (E) VE-cadherin, Tie1, Tie2, CD31, alpha-SMA, & fibronectin levels in HUVECs pre-incubated 16 h with 2% FBS-containing medium & then treated 24 h with PBS or 0.3 μg/ml of OPN. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29435158), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - miR-21-5p delivery by H-EV treatment promote NSCLC cell proliferation, survival & mobility by targeting PTEN, PDCD4 & RECK. a–d, western blot detecting PTEN, PDCD4 & RECK protein expression level in A549 or H23 cells after different treatment as described in Fig. 4. e–k, cell functional assay as described in Fig. 1. A549 or H23 cells with or without PTEN, PDCD4 or RECK gene overexpression (O/E) were tested. Wild type, non-treated A549 or H23 cells were used as negative control (NC). Bar in K indicates 20 μm. Tukey’s test was used for statistical analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30736829), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Expression of CTGF & TGF‐ beta1 in WT & MMP‐13 KO mice treated without & with NDMA. (A) Real‐time PCR analysis for the quantitative expression of CTGF, TGF‐ beta1, alpha‐SMA & collagen type 1 mRNA in WT & MMP‐13 KO mice. The data are mean ± S.D. of six samples per group. ***P < 0.001 NDMA‐treated WT mice versus control untreated WT mice; *P < 0.05 NDMA‐treated MMP‐13 KO mice versus untreated MMP‐13 KO mice; #P < 0.001 NDMA‐treated MMP‐13 KO mice versus NDMA‐treated WT mice. (B) Semiquantitative RT‐PCR for the expression of CTGF, TGF‐ beta1, alpha‐SMA & type 1 collagen mRNA in WT & MMP‐13 KO mice treated without & with NDMA. GAPDH was used as a loading control. The data are representative of six samples in each group. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28782260), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Hydrogen sulfide donors leads to inhibition of phosphate transport. Cells were cultured in a normal or calcific environment exposed to AP72 (20 μmol·L−1) for 5 days. (a) Phosphate content was determined using QuantiChrom quantitative colorimetric assay. (b) Pit1 & (c) Pit2 western blotting was performed & normalized to GAPDH. Data shown are means ± SEM of five independent experiments. *P < .05, significantly different as indicated; ns, not significant Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31017307), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Overexpression of NICD increases the total Fmod protein contents in BMDMs & LPS prevents the cleavage of Fmod in a dose dependent manner. (A,B) WB shows the expression of total Fmod in Apoe−/− & Notch1+/−;Apoe−/− BMDMs & quantification of three replicates as determined by Image J. (C,D) WB showing the total Fmod protein content in WT BMDMs 48 h post transfection with empty or NICD plasmids & the quantification of the immunoblots. (E,F) qRT-PCR showing the panel of M1 & M2 genes dysregulated with human recombinant FMOD (400 ng/ml for 24 h). (G) Co-immunoprecipitation showing contents of Tgf-beta 2, Fmod & NICD proteins pulled down with Fmod antibody from the WT & Notch1+/− peritoneal macrophages. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31142802), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Lactate dehydrogenase (LDH) assay & the Western blot of kidney injury molecule (KIM)-1 & N-cadherin in rat mesangial cells (RMCs). Cells were treated with bicalutamide (Bic) at concentrations of 30 & 60 μM at 24 h. (a) LDH release was increased by Bic (n = 3, * p < 0.05). (b) A representative blot of protein expressions of KIM-1 & N-cadherin. GAPDH was used as an internal control. (c) Quantitative data of Western blotting of KIM-1(n = 3, * p < 0.05). (d) Quantitative data of Western blotting of N-cadherin (n = 3, * p < 0.05). When RMCs were treated with Bic, N-cadherin dose-dependently decreased, however KIM-1 was significantly induced in the group treated with 60 μM. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32403414), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Immediate early gene (IEG) expression in RICH2 KO mice. IEG expression was analyzed in nuclear protein lysate from amygdala & normalized to GAPDH levels. No significant difference was detected in the expression of (A) activity-regulated cytoskeleton-associated protein (ARC; Unpaired t-test, n = 3, p = 0.077), & (B) c-FOS (Unpaired t-test, n = 3, p = 0.8784). A significant difference was detected in the expression of (C) cyclic AMP response element-binding protein (CREB; Unpaired t-test, n = 3, p = 0.002). The amount of phosphorylated CREB (pCREB) was unchanged (Unpaired t-test, n = 3, p = 0.1822). Exemplary Western blot bands are shown. Histone H3 was used a control for the presence of nuclear proteins in the lysate. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Relative Myonectin/CTRP15 content.Relative Myonectin/CTRP15 content was examined in the rat diaphragm muscle of the male lean zucker (LZR) & obese zucker rats (OZR). Exercised animals were trained on a motorized treadmill for 9 wk. Control animals were exposed to the similar environment (positioned next to the treadmill) but were not exercised. (A) Shows representative western blots for Myonectin & GAPDH. (B) The data are expressed in arbitrary units with values normalized to mean control value within phenotype. Image collected & cropped by CiteAb from the following publication (https://peerj.com/articles/605), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - FerrylHb induces HO-1 & ferritin in EC. Confluent HUVECs grown on 6-well plates were exposed to heme, Hb, metHb, & ferrylHb (100 μmol/L heme or as indicated in complete medium containing 15% of FBS). After 4 hours of incubation total RNA was isolated & HO-1 mRNA level was measured by quantitative RT-PCR (panels (a) & (d)). For protein expression, HUVECs were solubilized after 8 hours of treatment. HO-1 & ferritin H & L expression was detected by Western blot ((b) & (e)) or with ELISA (c). Immunoblots were reprobed with GAPDH & are representative of three independent experiments. Results are shown as mean ± S.D. (n = 3) from one representative experiment of three. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23766856), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Overexpression of NICD increases the total Fmod protein contents in BMDMs & LPS prevents the cleavage of Fmod in a dose dependent manner. (A,B) WB shows the expression of total Fmod in Apoe−/− & Notch1+/−;Apoe−/− BMDMs & quantification of three replicates as determined by Image J. (C,D) WB showing the total Fmod protein content in WT BMDMs 48 h post transfection with empty or NICD plasmids & the quantification of the immunoblots. (E,F) qRT-PCR showing the panel of M1 & M2 genes dysregulated with human recombinant FMOD (400 ng/ml for 24 h). (G) Co-immunoprecipitation showing contents of Tgf-beta 2, Fmod & NICD proteins pulled down with Fmod antibody from the WT & Notch1+/− peritoneal macrophages. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31142802), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Immediate early gene (IEG) expression in RICH2 KO mice. IEG expression was analyzed in nuclear protein lysate from amygdala & normalized to GAPDH levels. No significant difference was detected in the expression of (A) activity-regulated cytoskeleton-associated protein (ARC; Unpaired t-test, n = 3, p = 0.077), & (B) c-FOS (Unpaired t-test, n = 3, p = 0.8784). A significant difference was detected in the expression of (C) cyclic AMP response element-binding protein (CREB; Unpaired t-test, n = 3, p = 0.002). The amount of phosphorylated CREB (pCREB) was unchanged (Unpaired t-test, n = 3, p = 0.1822). Exemplary Western blot bands are shown. Histone H3 was used a control for the presence of nuclear proteins in the lysate. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - microRNA-200c regulates MARCKS.(A) The effect of miR-200c transfection on mRNA levels of predicted target genes in Wm266.4 melanoma cells was assessed using GAPDH as a control. Lower panel: immunoblot of MARCKS expression. (B) Brightfield imaging & morphological quantification of Wm266.4 cells transfected with siRNAs targeting miR-200c-responsive genes. (C) MARCKS knock-down by four individual siRNAs leads to rounded cell shape in Wm266.4 cells. (D) Levels of MARCKS mRNA in melanoma cell lines & normal melanocytes, measured by qPCR & normalized to levels of GAPDH. Error bars represent ± SEM; unpaired t-test *p<0.05 **p<0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20957176), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - microRNA-200c regulates MARCKS.(A) The effect of miR-200c transfection on mRNA levels of predicted target genes in Wm266.4 melanoma cells was assessed using GAPDH as a control. Lower panel: immunoblot of MARCKS expression. (B) Brightfield imaging & morphological quantification of Wm266.4 cells transfected with siRNAs targeting miR-200c-responsive genes. (C) MARCKS knock-down by four individual siRNAs leads to rounded cell shape in Wm266.4 cells. (D) Levels of MARCKS mRNA in melanoma cell lines & normal melanocytes, measured by qPCR & normalized to levels of GAPDH. Error bars represent ± SEM; unpaired t-test *p<0.05 **p<0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20957176), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Dose-dependent effects of LPS/IFN-gamma on Tgf-beta expression are associated with changes in the cleaved Fmod fragments. (A) Representative WB image showing the contents of NICD, Mmp8, total Fmod, cleaved Fmod, Tgf-beta 2 & Gapdh in macrophages in response to increasing dose of LPS (0, 5, 10, 25, 50 or 100 ng/ml) for 24 h. (B–F) Quantitation of immunoblots for NICD, Mmp8, total Fmod, cleaved Fmod, Tgf-beta 2 respectively (average of three replicates shown after normalizing the intensity with Gapdh. (WB = Western blot). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31142802), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Immediate early gene (IEG) expression in RICH2 KO mice. IEG expression was analyzed in nuclear protein lysate from amygdala & normalized to GAPDH levels. No significant difference was detected in the expression of (A) activity-regulated cytoskeleton-associated protein (ARC; Unpaired t-test, n = 3, p = 0.077), & (B) c-FOS (Unpaired t-test, n = 3, p = 0.8784). A significant difference was detected in the expression of (C) cyclic AMP response element-binding protein (CREB; Unpaired t-test, n = 3, p = 0.002). The amount of phosphorylated CREB (pCREB) was unchanged (Unpaired t-test, n = 3, p = 0.1822). Exemplary Western blot bands are shown. Histone H3 was used a control for the presence of nuclear proteins in the lysate. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - OPN induces EndoMT. (C) mRNA levels of VE-cadherin, Tie1, Tie2, CD31, alpha-SMA, & fibronectin in HUVECs pre-incubated 16 h with 2% FBS-containing medium & then treated 15 h with PBS or 0.3 μg/ml of OPN. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29435158), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - miR-200a associated elongation is associated with decreased actomyosin contractility.(A) The effect of miR-200a transfection on mRNA levels was measured for predicted target genes in Wm266.4 melanoma cells using GAPDH as a control. (B) Transfection of Wm266.4 cells with miR-200a leads to decreased pMLC levels. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20957176), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Regulation of cell migration-associated proteins by NCKU-21 in A549 & CL1-5 cells.Protein expressions of matrix metalloproteinase-2 (MMP-2) & MMP-9 were analyzed in A549 (A) & CL1-5 (B) cells treated with NCKU-21 for 24 hr. * P < 0.05 & ** P < 0.01, compared to the control group (without NCKU-21 treatment). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28945763), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Regulation of cell migration-associated proteins by NCKU-21 in A549 & CL1-5 cells.Protein expressions of matrix metalloproteinase-2 (MMP-2) & MMP-9 were analyzed in A549 (A) & CL1-5 (B) cells treated with NCKU-21 for 24 hr. * P < 0.05 & ** P < 0.01, compared to the control group (without NCKU-21 treatment). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28945763), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - TNF alpha negatively regulates EC differentiation: the effect of TNF alpha on VEGF-A-stimulated differentiation of MSCs into ECs was examined. TNF alphaR & VEGFR-2 mRNA expression was analyzed by RT-PCR (n = 3) (a). Sox18 protein levels were measured by Western blot analysis (n = 3) (b). Expression of EC markers was determined by FACS analysis, & a representative grid is shown (n = 3-4) ((c)–(g)). Endothelial tube formation was examined using an angiogenesis assay (n = 3) ((d)–(h)). Experiments were performed with samples taken from independent BM-MSC cultures from separate microswine. HUVECs were excluded from statistical analyses. Data are shown as mean ± SD. ∗p < 0.05 versus naïve MSCs & #p < 0.05 MSCs treated with VEGF-A versus VEGF-A plus TNF alpha cotreatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26106428), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - RICH2 is expressed at glutamatergic synapses in the amygdala. (A) Expression of Rich2 was detected on mRNA level using a PCR approach in amygdala using lysate from wild type (WT) mice. Cortex lysate was used as control as expression of Rich2 has been reported in cortex previously. The expression levels of Rich2 in amygdala were slightly higher compared to cortex. The primers used for amplification of Rich2 were placed in a region deleted in the genome of RICH2 Knockout (KO) mice. Absence of Rich2 expression in amygdala of RICH2 KO mice was confirmed. (B)In situ hybridization confirms Rich2 expression in the amygdala in adult WT animals. (C) RICH2 protein was not detected in RICH2 KO mice, but in amygdala of WT mice, & hippocampus & cortex as reported previously. ARP1 is a protein enriched in amygdala & was used as control to ensure proper preparation of amygdala tissue. (D) Immunocytochemistry performed on brain sections of WT mice reveals RICH2 signals co-localizing with HOMER1, a marker for excitatory postsynapses, in the amygdala. Top left: overview indicating magnified area in bottom left & right images. Arrows indicate clearly co-localizing signals (scale bar = 20 μm). Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - The absence of primary cilia in Kif3a null MEFs has a small effect on hyperosmotic induction of TonEBP without influencing its TAD activity (a) TonEBP/Nfat5 gene expression in wild-type & Kif3a null (Kif3a−/−) MEFs under hyper- (600 mOsm/kg H2O) & hypoosmotic (200 mOsm/kg H2O) conditions. (b,c) Western blot image & corresponding densitometry analyses showing TonEBP levels under different osmotic conditions in wild-type & Kif3a null MEFs. Kif3a null MEFs show slightly attenuated hyperosmotic increase but unaffected hypoosmotic decrease in TonEBP expression. (d,e) TonEBP-TAD (Ton-TAD) activity in wild-type & Kif3a null MEFs under hyperosmotic (d) & hypoosmotic (e) conditions (n = 4). Similar to wild-type cells, Kif3a null MEFs show increase & decrease in TonEBP-TAD activity under hyperosmotic & hypoosmotic conditions, respectively (n ≥ 3 with 3 technical replicates per biological replicate). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. Western blot images were cropped & acquired under same experimental conditions. See Supplementary Figs S1 & 3 for un-cropped Western blot images. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31664118), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Inhibition of AKT/GSK3 beta/ beta-catenin signaling pathway decreased HMGB1-induced EMT.(a) BEAS-2B cells treated w/ 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated w/ HMGB1 (300 ng/mL) for 24 h. Protein expression detected by WB analysis. Quantification of E-cadherin, ZO-1, & vimentin performed using ImageJ software. Data are expressed as mean ± SD (n = 5). #p < 0.05, as compared w/ control group. *p < 0.05, as compared w/ HMGB1-treated group. (b) BEAS-2B cells transduced w/ lentiviral-expressed beta-catenin shRNA (1 MOI) for 72 h & selected by puromycin. The mRNA from stable clones expressing beta-catenin-targeting shRNAs analyzed by quantitative real-time PCR. Data are expressed as mean ± SD (n = 5). ##p < 0.01, ###p < 0.001, as compared w/ control group. **p < 0.01, ***p < 0.001, as compared w/ vector group. (c) beta-catenin protein expression detected by WB analysis in a stable beta-catenin shRNA BEAS-2B cell clone. Quantification of protein expression performed using ImageJ software. Data are expressed as mean ± SD (n = 3). ##p < 0.01, ###p < 0.001, as compared w/ control group. **p < 0.01, ***p < 0.001, as compared w/ vector group. (d) A stable beta-catenin shRNA BEAS-2B cell clone treated w/ HMGB1 (300 ng/mL) for 24 h, & protein expression of E-cadherin, ZO-1, & vimentin detected by WB analysis. Quantification of protein expression performed using ImageJ software. Data are expressed as mean ± SD (n = 4). #p < 0.05, as compared w/ control group. *p < 0.05, as compared w/ HMGB1 group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots incubated w/ different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Ang II counteracts IL-6 or TNF alpha inhibition of EC differentiation: the combined effects of Ang II, IL-6, & TNF alpha on VEGF-A-stimulated differentiation of MSCs into ECs were examined. TNF alphaR, IL-6R, AT2R, & VEGFR-2 mRNA expression was analyzed by RT-PCR (n = 3) (a). Sox18 protein levels were measured by Western blot analysis (n = 3) (b). Expression of EC markers was determined by FACS analysis, & a representative grid is shown (n = 3-4) ((c)–(h)). Endothelial tube formation was examined using an angiogenesis assay (n = 3) ((d)–(i)). Experiments were performed with samples taken from independent BM-MSC cultures from separate microswine. HUVECs were excluded from statistical analyses. Data are shown as mean ± SD. ∗p < 0.05 versus naïve MSCs, #p < 0.05 MSCs treated with VEGF-A versus VEGF-A plus Ang II cotreatment, & alphap < 0.05 versus MSCs treated with VEGF-A & Ang II versus VEGF-A & Ang II plus IL-6 and/or TNF alpha. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26106428), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Inhibition of AKT/GSK3 beta/ beta-catenin signaling pathway decreased HMGB1-induced EMT.(a) BEAS-2B cells treated w/ 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated w/ HMGB1 (300 ng/mL) for 24 h. Protein expression detected by WB analysis. Quantification of E-cadherin, ZO-1, & vimentin performed using ImageJ software. Data are expressed as mean ± SD (n = 5). #p < 0.05, as compared w/ control group. *p < 0.05, as compared w/ HMGB1-treated group. (b) BEAS-2B cells transduced w/ lentiviral-expressed beta-catenin shRNA (1 MOI) for 72 h & selected by puromycin. The mRNA from stable clones expressing beta-catenin-targeting shRNAs analyzed by quantitative real-time PCR. Data are expressed as mean ± SD (n = 5). ##p < 0.01, ###p < 0.001, as compared w/ control group. **p < 0.01, ***p < 0.001, as compared w/ vector group. (c) beta-catenin protein expression detected by WB analysis in a stable beta-catenin shRNA BEAS-2B cell clone. Quantification of protein expression performed using ImageJ software. Data are expressed as mean ± SD (n = 3). ##p < 0.01, ###p < 0.001, as compared w/ control group. **p < 0.01, ***p < 0.001, as compared w/ vector group. (d) A stable beta-catenin shRNA BEAS-2B cell clone treated w/ HMGB1 (300 ng/mL) for 24 h, & protein expression of E-cadherin, ZO-1, & vimentin detected by WB analysis. Quantification of protein expression performed using ImageJ software. Data are expressed as mean ± SD (n = 4). #p < 0.05, as compared w/ control group. *p < 0.05, as compared w/ HMGB1 group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots incubated w/ different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HMGB1-activated AKT/GSK3 beta/ beta-catenin signaling pathways.(a) BEAS-2B cells were treated with different doses of HMGB1 for 10 min & expression of different proteins was detected by western blot. (b) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for expression of different proteins by western blot. (c) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin expression by western blot. (d) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin mRNA expression by real-time quantitative PCR. (e) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for nuclear & cytosolic beta-catenin expression by western blot. (f) BEAS-2B cells were treated with 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated with HMGB1 (300 ng/mL) for 24 h. beta-catenin expression was assessed by western blot analysis. Quantification of protein expression was performed using ImageJ software. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, as compared with the control group. #p < 0.05, as compared the HMGB1-treatment group. (A,B) (n = 4); (C–F) (n = 3). (g) Immunofluorescence staining of beta-catenin (green) was detected by fluorescence microscopy. Data are expressed as mean ± SD (n = 4). *p < 0.05, as compared with the 0-min group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots were incubated with different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Silencing of endogenous VEGF-A lacks an effect on mesenchymal stem cell differentiation into endothelial cells. Differentiating mesenchymal stem cells (MSCs) were transfected with small interfering RNA (siRNA) directed against vascular endothelial growth factor (VEGF-A) & then examined for endothelial cell (EC) marker expression. Treatment of MSCs with the cocktail of VEGF-A (2 ng/ml) plus angiotensin II (Ang II; 2 ng/ml) for 24 hours induced a synergistic increase in VEGF transcript as compared with VEGF-A alone or Ang II alone (A). VEGF-A mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (A). Standardization experiments demonstrated that 50 nM siRNA directed against VEGF completely abrogated VEGF protein expression (B). Fluorescence-activated cell sorting analysis revealed that there was no significant difference in EC marker expression between differentiating MSCs that received VEGF-A siRNA or scrambled control siRNA (C, D, E). Note that human umbilical vein endothelial cells (HUVECs) were excluded from the statistical analyses. *P <0.05 vs. naïve MSCs. #P <0.05 vs. VEGF-A (2 ng/ml)-treated MSCs or Ang II (2 ng/ml)-treated MSCs alone, n = 3. Image collected & cropped by CiteAb from the following publication (https://stemcellres.biomedcentral.com/articles/10.1186/scrt538), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - N-EV & H-EV treatment promote macrophage M2 polarization by delivering miR-21-5p that targets PTEN. a, western blot analysis of PTEN protein expression level in induced macrophages. H/i-miR-EV, monocytes were induced with the presence of EV secreted by miR-21-5p-inhibited, hypoxia pre-challenged MSCs; H-EV + i-miR, monocytes were transfected with miR-21-5p inhibitor-expressing vector before induction with the presence of H-EV. Macrophages induced without MSC-EV were used as negative control (NC). b, c, flow cytometry determining the percentage of CD163+CD206+ cells among total CD68+ cells after induction. N-EV + O/E PTEN or H-EV + O/E PTEN, monocytes were transfected with PTEN overexpressing vector before N-EV or H-EV treatment, respectively. d–f, western blot detecting Akt & STAT3 protein expression as well as their activating phosphorylation (p-Ser473 for Akt & p-tyr705 for STAT3) in macrophages after induction. g–i, ELISA evaluating IL-10, TGF-beta & VEGF-alpha in macrophage culture medium after induction. Macrophages induced with the presence of N-EV were used as negative control in b–i. Tukey’s test was used for statistical analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30736829), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Demonstration of the advantages of crude fractionation over whole cell lysis & scale up of the approach. (A) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well plate & subsequently lysed directly in NP40 or RIPA lysis buffer or processed according to the protocol in Figure 2 (Sequential). An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by staining with Coomassie blue or by Western blotting & probing with antibodies to various marker proteins of specific intracellular organelles as previously. (B) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish & then transfected the next day in triplicate with 400 ng of a construct encoding V5/6×-HIS tagged ERGIC-53. Cells were extracted 48 hours later either directly in RIPA buffer, directly in NP40 buffer or according to our extraction protocol. Proteins were then partially purified from each extract using Nickel resin (SIGMA #P6611) & eluted proteins were analyzed by 10% SDS PAGE followed by staining with Coomassie blue or by Western blotting & probing with the anti V5 antibody. (C) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish & then transfected the next day in with 1600 ng of pcDNA6/V5-His (Invitrogen) or constructs encoding WT or MUT COMP. Cells were harvested 48 hours later & extracted according to our protocol. (D) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish or 8.4 × 105 cells per well of a 35 mm dish & processed according to the protocol in Figure 2. An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by Western blotting & probing with antibodies to GAPDH, BiP & Lamin A. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20003239), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Notch1, ApoER2 & Dab1 colocalize in hippocampal neurons. (A) Representative image of fluorescent immunolabeling shows that Notch1 is localized in a subset of CA3 neurons, co-expressing Dab1 & ApoER2 (magnified insert). ApoER2 is highly expressed in all CA3 neurons. (B) Fluorescent immunolabeling shows the distribution of Notch1 & Dab1 as compared to Arc/Arg3.1 in CA3 neurons. (B′) Close up of a dendrite showing colocalization of the three proteins (white arrowheads). (C) Western blot analysis on immunoprecipitated samples from whole hippocampal lysate shows that Notch1 displays a strong interaction with Dab1 whereas Notch1 & ApoER2 interaction appears weaker in the preparation (n = 4 independent experiments). (D) Immunolabeling on immunoprecipitated samples from whole cortical lysates show that Notch1 & Dab1 interact. ApoER2 is bound to both Notch1 & Dab1 (n = 3 independent experiments). (E) Co-IP from synaptosomal membrane fractions reveals a stronger interaction between Notch1 & ApoER2, which is reduced in the N1cKO (n = 3 independent pulldowns). In (C–E) GAPDH is used as a loading control for the inputs & to detect contamination in the IP samples. Scale bar in (A) is 50 μm & (B) 25 μm. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26635527), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Reduced expression of UNC5A in FancC−/− cells. a Reduced expression of Unc5A in brain cortex of FancC−/− mice compared to wild-type littermates. Western blotting was performed with the indicated antibodies. Each lane represents a different animal (WT: n = 6; FancC−/−: n = 8). b Bar graph represents ratio of Unc5A normalized to tubulin from WT (n = 6) & FancC−/− mice (n = 8). c Bar graph representation of UNC5A expression normalized to Sdha & Tbp from wild-type (WT) & FancC−/−-derived fibroblasts (n = 3). RLU: Relative light units. d–f HEK293T cells were transfected with UNC5AICD, FANCE & FANCC constructs expressing full-length FANCC (in d), FANCC1–306 (in e) or FANCC307–558 (in f). Constructs were transfected at a molecular ratio of 1:1:1 or with 10 times the molar amount of FANCE, FANCC or UNC5AICD as indicated in the figure. The total amount of transfected plasmid was equalized for all strategies with control empty vectors. Representative immunoblots performed with anti-HA (UNC5AICD), anti-FANCC, anti-FANCE or anti-GAPDH antibodies are shown. Arrows indicate appropriate protein bands. Bar graphs represent the mean fold change ± SEM of UNC5A protein levels normalized to GAPDH compared to 1:1:1 transfection controls in at least 4 separate experiments. *p < 0.05; **p < 0.005 Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30213274), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Analysis of GFAP in mouse models of AxD by immunoblotting.Urea-soluble fractions extracted from wild type (GFAP+/+), R236H mutant knockin (GFAPR236H/+) & human wild type GFAP transgenic (GFAPTg) mice (n = 3 for each genotype) were analyzed by immunoblotting with indicated anti-GFAP antibodies (A-D). Immunoblots were also probed with antibodies specific to alphaB-crystallin (F) & GAPDH (E), which was used as a loading control. Representative images were shown, with the antibody used for immunoblotting was indicated above each panel. Approximate molecular weight markers (in kDa) were shown on the right. Dashed lines indicated lanes that samples run on the same gel but were noncontiguous (D & F). Notice that although all anti-GFAP antibodies tested recognized full-length GFAP (A-D), both 2.2B10 (A) & SMI-21 (B) antibodies also detected GFAP degradation products sized between 25 & 35 kDa in GFAPTg mice. The level of alphaB-crystallin (F) was increased in both GFAPR236H/+ & GFAPTg mice compared to wild type controls (GFAP+/+). Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0180694), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Knockdown of Ift88 or Kif3a in NP cells does not affect hyperosmotic upregulation of TonEBP. (a) TonEBP/Nfat5 mRNA levels in NP cells with Ift88 knockdown (n ≥ 5). (b) Western blot image showing increased TonEBP expression in response to hyperosmolarity (550 mOsm/kg H2O) independently of Ift88 knockdown. (c) Densitometry analyses of TonEBP with Ift88 knockdown (n ≥ 4). (d) TonEBP/Nfat5 mRNA levels in NP cells with Kif3a knockdown (n ≥ 3). (e) Western blot image showing that hyperosmotic induction of TonEBP is maintained after Kif3a knockdown. (f) Densitometry analyses of TonEBP after Kif3a knockdown (n ≥ 4). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. Western blot images were cropped & acquired under same experimental conditions. See Supplementary Fig. S1-1 for un-cropped Western blot images. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31664118), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - S166 phosphorylation facilitates necrosome formation.a BMDMs from mice of the indicated genotypes were treated with TNF (T; 10 ng ml−1), SMAC mimetic (S (Birinapant); 1 µM) & Z-VAD-FMK (Z) for 0, 2 & 4 h. RIPK1 immunoprecipitates & total cell lysates (input) were analyzed by immunoblotting with the indicated antibodies. b BMDMs from mice of the indicated genotypes were treated with TNF (T; 10 ng ml−1), SMAC mimetic (S (Birinapant); 1 µM) & Z-VAD-FMK (Z) for 2 & 4 h. Lysates were processed under non-reducing conditions & analyzed for presence of MLKL by immunoblot. c BMDMs from mice of the indicated genotypes were treated with LPS (L; 100 ng ml−1) & Z- VAD-FMK (Z) for 13 & 15 h. Lysates were processed under non-reducing or reducing conditions & analyzed by immunoblot using the indicated antibodies. d BMDMs from mice of the indicated genotypes were treated with Poly(I:C) (P; 0.5 μg ml−1) & Z-VAD-FMK (Z) for 13 & 15 h. Lysates were processed under non-reducing or reducing conditions & analyzed by immunoblot using the indicated antibodies. a, c Representative data of three independent experiments is shown. b, d Representative data of two independent experiments is shown. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32269263), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Notch1 affects CREB phosphorylation & transcription. (A) Immunoblot on whole hippocampal lysates shows a substantial reduction in CamKII in hippocampi of N1cKO as compared to WT. beta-actin is used as a loading control (n = 3 independent experiments). (B) Quantification of CamKII protein expression between N1cKO & WT normalized to beta-actin shows a reduction by 78% in N1cKO hippocampi (n = 6 animals per genotype; Student’s t-test, p < 0.05). (C) Western blot on hippocampal lysates shows ERK1/2 phosphorylation (pERK1/2) in N1cKO is reduced as compared to WT hippocampi. beta-actin is used as a loading control (n = 3 independent experiments). (D) Quantification of ERK1/2 phosphorylation in N1cKO as compared to WT indicates a 76% decrease in KO samples (n = 6 mice per genotype; Student’s t-test, p < 0.05). (E) Immunoblot on whole hippocampal lysates shows a reduction in CREB phosphorylation in N1cKO as compared to WT (n = 3 independent experiments). (F) Bar graph shows a near to significance 56% reduction in CREB phosphorylation in N1cKO hippocampi as compared to WT (n = 4 animals per genotype; Student’s t-test, p = 0.069). (G) Graph summarizing transcripts expression of CREB targets, Egr1, Bdnf & c-fos, in hippocampi from cage control (CC) & environmental exposed (EE) WT & N1cKO mice (n = 3–4 mice per genotype; Student’s t-test, p < 0.05; Black bars compare WT CC w/ WT EE; gray bars compare WT EE w/ N1cKO EE). (H) Model of proposed Notch1 non-canonical signaling regulations of Reelin, NMDA & CREB pathways described in this study. Elements putatively connecting Notch1/Reelin signaling to Notch1/Glutamaterigic signaling are dotted & are based on existing literature. Data are averages ± SEM & *=p < 0.05; 0 signifies p = 0.5. Image collected & cropped by CiteAb from following publication (https://pubmed.ncbi.nlm.nih.gov/26635527), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Altered Rho GTPase signaling in amygdala of RICH2 KO mice. (A) Comparison between the GTPase activating protein (GAP) activity of the P2 lysates from WT & RICH2 KO mice. The GAP activity is measured in nmoles/min/mg of small G-protein. First a standard curve is made using the absorbance of KH2PO4 at different concentrations, later the absorbance of WT & RICH2 KO lysates are plotted on the graph. The GAP activity is higher for WT lysates compared to RICH2 KO lysates. (B) In the presence of the small G protein RhoA, the absorbance of WT lysates is higher than that of RICH2 KO lysate, which indicates that RICH2 is acting as a GAP for RhoA in amygdala. (C) There is no difference between the absorbance of WT & RICH2 KO lysates in the presence of RAC1 & CDC42 (D,E) Like RAC1 & CDC42, RAS p21 is not a target G protein for RICH2 in amygdala as there is no difference in the absorbance of WT & RICH2 KO lysates in the presence of RAS p21. (F) Western blot analysis of amygdala P2 lysates reveals that expression of RhoA is significantly higher in RICH2 KO lysates, in comparison with WT lysates (Unpaired t-test, p = 0.0508, n = 3). (G,H) Western blot analysis revealed no alteration in the expressions of Rac1 (G) & Cdc42 (H) between WT & RICH2 KO using amygdala P2 lysates. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Application of the approach to other cell lines. (A) HT1080 cells (2 × 105) & HeLa cells (1 × 105) were plated in the wells of a 12 well plate & the digitonin concentration required for optimal extraction of the cytosol was determined as in Figure1. (B) The remainder of the protocol from Figure 2 was then applied using 100 μg/ml digitonin as the optimal cytosolic extract in each case. An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by Western blotting & probing with antibodies to GAPDH, BiP & Histone H3 (Used as an alternative nuclear marker due to weakness of the Lamin A signal in HeLa cells). (C) Samples were analyzed by 4-12% SDS PAGE followed by staining with Coomassie blue & then silver staining showing the difference in protein composition of each sample. The bracket alongside the gels marks the position of Histones in the RIPA nuclear extract & the asterisk next to the silver stained gels denotes protein bands unique to the E-RIPA extracts. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20003239), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Optimization of digitonin concentration for the extraction of cytosolic proteins. (A) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well plate & were harvested 48 hours later. Cells were then lysed in 400 μl of buffer containing digitonin at the concentration indicated. Following centrifugation & collection of the supernatant the remaining cell pellet was further extracted in NP40 lysis buffer. An aliquot of each extract was then analysed by 4-12% SDS PAGE (Invitrogen #NP0322) followed by staining with Coomassie blue. (B) The concentration of protein in each extract was determined by colorimetric protein assay (Pierce #23232) & the results were plotted to demonstrate that total protein extracted was the same regardless of the starting concentration of digitonin. (C) Extracted proteins from each fraction were analyzed by Western blot using an anti GAPDH antibody (NOVUS Biologicals #300-221B) as a marker of the cytosol & an anti BiP antibody (SIGMA # G918) as a marker of the endoplasmic reticulum. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20003239), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - CCL2 promotes prostate cancer cell migration & induces CCL22 secretion, which is a ligand of CCR4A. Prostate cancer cells are placed in transwell inserts & treated with CCL2 (0–30 ng/ml). After 24-h incubation, PC-3 & DU145 cells that had migrated through the membrane are stained. The mean OD value is read using a microreader at 595 nm. Migration of LNCaP cells is assessed with a wound-healing assay. Data are presented as mean ± SD. B. DU145 cells are co-cultured with THP-1 & U937 cells & treated with CCL2 (30 ng/ml) for 24 h, CM is collected, & CCL17 & CCL22 levels are analyzed using ELISA. The mean OD value is read using a microreader at 450 nm, & data are presented as mean ± SD. C, D. Total RNA & protein are extracted from prostate cancer cells, & CCR2 & CCR4 gene & protein expression levels are analyzed using PCR (C) & western blot (D). E. Prostate cancer cells (1.0 × 105 cells/well) are seeded into 6-well plates & cultured until they reach 60%–70% confluence. Cells are incubated with anti-CCR2 or anti-CCR4 antibody & detected using a second antibody conjugated with FITC (green). Cells are counterstained with 4',6-diamidino-2-phenylindole (blue). Adjustments of brightness, contrast, & size are applied to the whole images of western blot-based analyses without elimination of any information present in the original, including backgrounds. All experiments are performed in triplicate, & mean values are shown. *p < 0.05, **p < 0.01, ***p < 0.001. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28039457), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Inhibition of AKT/GSK3 beta/ beta-catenin signaling pathway decreased HMGB1-induced EMT.(a) BEAS-2B cells treated w/ 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated w/ HMGB1 (300 ng/mL) for 24 h. Protein expression detected by WB analysis. Quantification of E-cadherin, ZO-1, & vimentin performed using ImageJ software. Data are expressed as mean ± SD (n = 5). #p < 0.05, as compared w/ control group. *p < 0.05, as compared w/ HMGB1-treated group. (b) BEAS-2B cells transduced w/ lentiviral-expressed beta-catenin shRNA (1 MOI) for 72 h & selected by puromycin. The mRNA from stable clones expressing beta-catenin-targeting shRNAs analyzed by quantitative real-time PCR. Data are expressed as mean ± SD (n = 5). ##p < 0.01, ###p < 0.001, as compared w/ control group. **p < 0.01, ***p < 0.001, as compared w/ vector group. (c) beta-catenin protein expression detected by WB analysis in a stable beta-catenin shRNA BEAS-2B cell clone. Quantification of protein expression performed using ImageJ software. Data are expressed as mean ± SD (n = 3). ##p < 0.01, ###p < 0.001, as compared w/ control group. **p < 0.01, ***p < 0.001, as compared w/ vector group. (d) A stable beta-catenin shRNA BEAS-2B cell clone treated w/ HMGB1 (300 ng/mL) for 24 h, & protein expression of E-cadherin, ZO-1, & vimentin detected by WB analysis. Quantification of protein expression performed using ImageJ software. Data are expressed as mean ± SD (n = 4). #p < 0.05, as compared w/ control group. *p < 0.05, as compared w/ HMGB1 group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots incubated w/ different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Stable knockdown of Ift88 or Kif3a inhibits formation of NP cell primary cilia. (a) Ift88 mRNA levels in NP cells transduced with control (ShCtr) or two different ShIft88 clones were measured by qRT-PCR to confirm the knockdown (n ≥ 5). (b) Western blot image showing significant reduction of IFT88 protein levels after the knockdown of Ift88. (c) Densitometry analyses of Western blots confirm significant knockdown of IFT88 (n ≥ 5). (d–f) qRT-PCR, Western blot, & corresponding densitometry analyses, show significant downregulation of KIF3A after stable knockdown using two different ShKif3a clones (n ≥ 4). (g) Acetylated alpha-tubulin immunofluorescence staining after lentiviral transduction of ShIft88 or ShKif3a shows inhibition of primary cilia formation in majority of rat NP cells. Scale bar = 75 μm. White arrowheads point to primary cilia. (h,i) Quantitation of percentage of NP cells with primary cilia & primary cilium length after stable silencing of Ift88 or Kif3a (n = 3; at least 150 cells/group). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s, Holm-Sidak’s, or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. For statistical comparison of the percentages of NP cells with primary cilia, Fisher’s exact test was used. Western blot images were cropped & acquired under same experimental conditions. See Supplementary Fig. S1–1 for un-cropped Western blot images. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31664118), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HMGB1-activated AKT/GSK3 beta/ beta-catenin signaling pathways.(a) BEAS-2B cells were treated with different doses of HMGB1 for 10 min & expression of different proteins was detected by western blot. (b) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for expression of different proteins by western blot. (c) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin expression by western blot. (d) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin mRNA expression by real-time quantitative PCR. (e) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for nuclear & cytosolic beta-catenin expression by western blot. (f) BEAS-2B cells were treated with 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated with HMGB1 (300 ng/mL) for 24 h. beta-catenin expression was assessed by western blot analysis. Quantification of protein expression was performed using ImageJ software. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, as compared with the control group. #p < 0.05, as compared the HMGB1-treatment group. (A,B) (n = 4); (C–F) (n = 3). (g) Immunofluorescence staining of beta-catenin (green) was detected by fluorescence microscopy. Data are expressed as mean ± SD (n = 4). *p < 0.05, as compared with the 0-min group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots were incubated with different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - CCL2 increases not only CCL22 but also CCR2 & CCR4A. Prostate cancer cells are treated with CCL2 (30 ng/ml) for 24 h, & western blot for CCR2 is performed. B. Prostate cancer cells are treated with CCL2, CCL17, & CCL22 (30 ng/ml) for 24 h, & western blot for CCR4 is performed. C. Prostate cancer cells are co-cultured with U937 or U937-M cells for 24 h, & western blot for CCR4 is performed. D. Prostate cancer cells are placed in transwell inserts & treated with CCL17 & CCL22 (0–30 ng/ml). After 24-h incubation, PC-3 & DU145 cells that had migrated through the membrane are stained. The mean optical density (OD) value is read using a microreader at 595 nm. Migration of LNCaP cells is assessed with a wound-healing assay. Adjustments of brightness, contrast, & size are applied to the whole images of western blot-based analyses without elimination of any information present in the original, including backgrounds. Data are presented as mean ± SD. All experiments are performed in triplicate, & mean values are shown. *p < 0.05, **p < 0.01, ***p < 0.001. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28039457), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - ATR2 siRNA transfection & immunophenotyping for EC markers. I Concentration selection for siRNA transfection. Three different concentrations (10, 35, & 50 nM) of ATR2 siRNA were used according to the manufacturer’s protocol. Western blot analysis showed inhibition of ATR2 by 10, 35, & 50 nM of ATR2 siRNA. However, 50 nM of ATR2 siRNA showed the highest inhibition among all three different concentrations (A). ATR2 silencing by siRNA transfection with EGM compared with AMSCs with EGM & EGM + scrambled siRNA (negative control) (B). GAPDH was used as a housekeeping gene. II Flow cytometric analysis of PECAM1 (CD31) in four different groups; control group with EGM (A), AMSCs with EGM & MMP-2 siRNA (B), AMSCs with EGM & MMP-14 siRNA (C), & HUVECs as the positive control (D). Cell transfection with 5 μM of ATR2 siRNA for EGM (E), AMSCs with EGM & MMP-2 siRNA (F), & AMSCs with EGM & MMP-14 siRNA (G). Flow cytometry data were analyzed to show the significant differences between the groups (H). III Flow cytometric analysis of VE-cadherin (CD144) in four different groups: control group AMSCs with EGM (A), AMSCs with EGM & MMP-2 siRNA (B), AMSCs with EGM & MMP-14 siRNA (C), & HUVECs as the positive control (D). Cell transfection with 5 μM of ATR2 siRNA for EGM (E), AMSCs with EGM & MMP-2 siRNA (F), & AMSCs with EGM & MMP-14 siRNA (G). Flow cytometry data were analyzed to show the significant differences between the groups (H). **p < 0.01. EBM endothelial cell basal medium, EGM endothelial cell growth medium, MMP matrix metalloproteinase, ATR2 angiotensin receptor R2, HUVEC human umbilical vein endothelial cell Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28499402), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Altered Rho GTPase signaling in amygdala of RICH2 KO mice. (A) Comparison between the GTPase activating protein (GAP) activity of the P2 lysates from WT & RICH2 KO mice. The GAP activity is measured in nmoles/min/mg of small G-protein. First a standard curve is made using the absorbance of KH2PO4 at different concentrations, later the absorbance of WT & RICH2 KO lysates are plotted on the graph. The GAP activity is higher for WT lysates compared to RICH2 KO lysates. (B) In the presence of the small G protein RhoA, the absorbance of WT lysates is higher than that of RICH2 KO lysate, which indicates that RICH2 is acting as a GAP for RhoA in amygdala. (C) There is no difference between the absorbance of WT & RICH2 KO lysates in the presence of RAC1 & CDC42 (D,E) Like RAC1 & CDC42, RAS p21 is not a target G protein for RICH2 in amygdala as there is no difference in the absorbance of WT & RICH2 KO lysates in the presence of RAS p21. (F) Western blot analysis of amygdala P2 lysates reveals that expression of RhoA is significantly higher in RICH2 KO lysates, in comparison with WT lysates (Unpaired t-test, p = 0.0508, n = 3). (G,H) Western blot analysis revealed no alteration in the expressions of Rac1 (G) & Cdc42 (H) between WT & RICH2 KO using amygdala P2 lysates. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Reduced expression of UNC5A in FancC−/− cells. a Reduced expression of Unc5A in brain cortex of FancC−/− mice compared to wild-type littermates. Western blotting was performed with the indicated antibodies. Each lane represents a different animal (WT: n = 6; FancC−/−: n = 8). b Bar graph represents ratio of Unc5A normalized to tubulin from WT (n = 6) & FancC−/− mice (n = 8). c Bar graph representation of UNC5A expression normalized to Sdha & Tbp from wild-type (WT) & FancC−/−-derived fibroblasts (n = 3). RLU: Relative light units. d–f HEK293T cells were transfected with UNC5AICD, FANCE & FANCC constructs expressing full-length FANCC (in d), FANCC1–306 (in e) or FANCC307–558 (in f). Constructs were transfected at a molecular ratio of 1:1:1 or with 10 times the molar amount of FANCE, FANCC or UNC5AICD as indicated in the figure. The total amount of transfected plasmid was equalized for all strategies with control empty vectors. Representative immunoblots performed with anti-HA (UNC5AICD), anti-FANCC, anti-FANCE or anti-GAPDH antibodies are shown. Arrows indicate appropriate protein bands. Bar graphs represent the mean fold change ± SEM of UNC5A protein levels normalized to GAPDH compared to 1:1:1 transfection controls in at least 4 separate experiments. *p < 0.05; **p < 0.005 Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30213274), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - N-EV & H-EV treatment promote macrophage M2 polarization by delivering miR-21-5p that targets PTEN. a, western blot analysis of PTEN protein expression level in induced macrophages. H/i-miR-EV, monocytes were induced with the presence of EV secreted by miR-21-5p-inhibited, hypoxia pre-challenged MSCs; H-EV + i-miR, monocytes were transfected with miR-21-5p inhibitor-expressing vector before induction with the presence of H-EV. Macrophages induced without MSC-EV were used as negative control (NC). b, c, flow cytometry determining the percentage of CD163+CD206+ cells among total CD68+ cells after induction. N-EV + O/E PTEN or H-EV + O/E PTEN, monocytes were transfected with PTEN overexpressing vector before N-EV or H-EV treatment, respectively. d–f, western blot detecting Akt & STAT3 protein expression as well as their activating phosphorylation (p-Ser473 for Akt & p-tyr705 for STAT3) in macrophages after induction. g–i, ELISA evaluating IL-10, TGF-beta & VEGF-alpha in macrophage culture medium after induction. Macrophages induced with the presence of N-EV were used as negative control in b–i. Tukey’s test was used for statistical analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30736829), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Protein expression of CTGF, TIMP‐1, TGF‐ beta1, alpha‐SMA & collagen type 1 in WT & MMP‐13 KO mice treated without & with NDMA. (A) Western blotting for the proteins of full‐length CTGF & its C‐terminal fragments, TIMP‐1, active TGF‐ beta1, alpha‐SMA & collagen type 1 in the liver tissues from WT & MMP‐13 KO mice. The images are representative of six Western blots per each group. (B) Quantitative analysis of Western blot images. The data are mean ± S.D. of six images per group. ***P < 0.001 NDMA‐treated WT mice versus control untreated WT mice & NDMA‐treated MMP‐13 KO mice versus untreated MMP‐13 KO mice; **P < 0.01 NDMA‐treated MMP‐13 KO mice versus untreated MMP‐13 KO mice; *P < 0.05 NDMA‐treated MMP‐13 KO mice versus untreated MMP‐13 KO mice; #P < 0.001 NDMA‐treated MMP‐13 KO mice versus NDMA‐treated WT mice; @, P < 0.05 NDMA‐treated MMP‐13 KO mice versus NDMA‐treated WT mice. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28782260), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Endothelial responses provoked by Hb-lipid interactions are attenuated by sulfide donors. (a) OxLDL (200 μg/mL) was incubated with sulfide donors NaSH, GYY4137, AP67, & AP72 at the concentrations of 20 & 200 μmol/L at 37°C for 24 hours. Confluence HAoECs were exposed to the LDL samples for 4 hours then cell viability was determined by MTT assay. Representative experiment, n = 3, each performed in 8 wells in parallel. (b) OxLDL (200 μg/mL) was incubated with sulfide donors NaSH, GYY4137, AP67, & AP72 at the concentration of 20 & 200 μmol/L at 37°C for 24 hours. Confluence HAoECs were exposed to LDL samples (50 μg/mL) for 8 hours. HO-1 & HO-2 protein expressions were determined by Western blotting. Representative experiment, n = 3. (c–e) Hb (10 μmol/L) was incubated with H2O2 (50 μmol/L) in the presence or absence of sulfide donors NaSH, GYY4137, AP67, & AP72 (200 μmol/L) at 37°C for 90 minutes. (c) Confluence HAoECs were exposed to the obtained Hb samples for 8 hours & VCAM-1 expression was determined by Western blotting. Representative experiment, n = 3. (d, lower panel) Confluence HAoECs cultured in ECIS plates were exposed to the obtained Hb samples & transendothelial electrical resistance was monitored by ECIS instrument for 3 hours. Representative experiment, n = 3, each performed in triplicate (d, upper panel). HAoECs grown on coverslips, upon reaching confluence cells were exposed to the Hb samples for 8 hours. Cells were stained for F-actin (phalloidin-TRITC, red) & DNA (Hoechst, blue). White arrows show intercellular gaps. Representative image, n = 5. ∗∗ indicates statistical significance (p < 0.001). ∗∗∗ indicates statistical significance (p < 0.0001). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29560080), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Stable knockdown of Ift88 or Kif3a inhibits formation of NP cell primary cilia. (a) Ift88 mRNA levels in NP cells transduced with control (ShCtr) or two different ShIft88 clones were measured by qRT-PCR to confirm the knockdown (n ≥ 5). (b) Western blot image showing significant reduction of IFT88 protein levels after the knockdown of Ift88. (c) Densitometry analyses of Western blots confirm significant knockdown of IFT88 (n ≥ 5). (d–f) qRT-PCR, Western blot, & corresponding densitometry analyses, show significant downregulation of KIF3A after stable knockdown using two different ShKif3a clones (n ≥ 4). (g) Acetylated alpha-tubulin immunofluorescence staining after lentiviral transduction of ShIft88 or ShKif3a shows inhibition of primary cilia formation in majority of rat NP cells. Scale bar = 75 μm. White arrowheads point to primary cilia. (h,i) Quantitation of percentage of NP cells with primary cilia & primary cilium length after stable silencing of Ift88 or Kif3a (n = 3; at least 150 cells/group). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s, Holm-Sidak’s, or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. For statistical comparison of the percentages of NP cells with primary cilia, Fisher’s exact test was used. Western blot images were cropped & acquired under same experimental conditions. See Supplementary Fig. S1–1 for un-cropped Western blot images. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31664118), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Upregulation & accumulation of the endogenous mouse GFAP in the GFAPTg mice.Urea-soluble fractions prepared from whole brains of GFAP+/+ wild type (A, lanes 1–3) & GFAPTg (A, lanes 4–6) mice were analyzed by immunoblotting using the anti-mouse GFAP antibody. Immunoblots probed with anti-GAPDH antibody was used as a loading control. Note that the endogenous mouse GFAP was significantly increased in the GFAPTg mice (A, lanes 4–6) compared to wild type controls (A, lanes 1–3). Approximate molecular weight markers (in kDa) were shown on the left. The distributions of human GFAP in relation to the endogenous mouse GFAP in brain sections of wild type (GFAP+/+, B) & GFAPTg (C-H) mice were visualized by double-label immunofluorescence microscopy using anti-mouse GFAP & SMI-21 antibodies. The immunofluorescence for human GFAP was in the green channel (C & F), whereas the counterstaining for mouse GFAP was in the red channel (D & G). Merged images showed the superimposition of both the green & red signals, with overlapping area appearing yellow (B, E, & H). Nuclei were revealed by staining with DAPI (B, E, & H). In wild type mice, normal appearance of GFAP in astrocytes was readily stained with the anti-mouse GFAP antibody (B), with very little staining with the SMI-21 antibody. Conversely, GFAPTg mice showed numerous Rosenthal fiber-like GFAP aggregates in the granular cell layer of the cerebellum (C-E) & the pial surface of the cortex (F-H), which were immunopositive for both mouse & human GFAP. Bar, 10 μm. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0180694), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - IL-6 negatively regulates EC differentiation: the effect of IL-6 on VEGF-A-stimulated differentiation of MSCs into ECs was examined. IL-6R & VEGFR-2 mRNA expression was analyzed by RT-PCR & normalized to GAPDH (n = 3) (a). Sox18 protein levels were measured by Western blot analysis & normalized to GAPDH (n = 3) (b). Expression of EC markers was determined by FACS analysis, & a representative grid is shown (n = 3–6) ((c)–(g)). Endothelial tube formation was examined using an angiogenesis assay (n = 3) ((d)–(h)). Experiments were performed with samples taken from independent BM-MSC cultures from separate microswine. HUVECs were excluded from statistical analyses. Data are shown as mean ± SD. ∗p < 0.05 versus naïve MSCs & #p < 0.05 MSCs treated with VEGF-A versus VEGF-A plus IL-6 cotreatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26106428), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Demonstration of the advantages of crude fractionation over whole cell lysis & scale up of the approach. (A) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well plate & subsequently lysed directly in NP40 or RIPA lysis buffer or processed according to the protocol in Figure 2 (Sequential). An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by staining with Coomassie blue or by Western blotting & probing with antibodies to various marker proteins of specific intracellular organelles as previously. (B) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish & then transfected the next day in triplicate with 400 ng of a construct encoding V5/6×-HIS tagged ERGIC-53. Cells were extracted 48 hours later either directly in RIPA buffer, directly in NP40 buffer or according to our extraction protocol. Proteins were then partially purified from each extract using Nickel resin (SIGMA #P6611) & eluted proteins were analyzed by 10% SDS PAGE followed by staining with Coomassie blue or by Western blotting & probing with the anti V5 antibody. (C) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish & then transfected the next day in with 1600 ng of pcDNA6/V5-His (Invitrogen) or constructs encoding WT or MUT COMP. Cells were harvested 48 hours later & extracted according to our protocol. (D) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish or 8.4 × 105 cells per well of a 35 mm dish & processed according to the protocol in Figure 2. An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by Western blotting & probing with antibodies to GAPDH, BiP & Lamin A. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20003239), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HMGB1-activated AKT/GSK3 beta/ beta-catenin signaling pathways.(a) BEAS-2B cells were treated with different doses of HMGB1 for 10 min & expression of different proteins was detected by western blot. (b) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for expression of different proteins by western blot. (c) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin expression by western blot. (d) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin mRNA expression by real-time quantitative PCR. (e) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for nuclear & cytosolic beta-catenin expression by western blot. (f) BEAS-2B cells were treated with 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated with HMGB1 (300 ng/mL) for 24 h. beta-catenin expression was assessed by western blot analysis. Quantification of protein expression was performed using ImageJ software. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, as compared with the control group. #p < 0.05, as compared the HMGB1-treatment group. (A,B) (n = 4); (C–F) (n = 3). (g) Immunofluorescence staining of beta-catenin (green) was detected by fluorescence microscopy. Data are expressed as mean ± SD (n = 4). *p < 0.05, as compared with the 0-min group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots were incubated with different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Co-culture of macrophages & prostate cancer cells increases prostate cancer cell migration & invasion & induces CCL2 secretionA. U937 differentiation to M2 macrophages is determined using western blot analysis. CCR7 (an M1-type macrophage marker) & CD206 (an M2-type macrophage marker) are assayed using proteins extracted from U937 cells treated with PMA for 24 h & exposed to CM of prostate cancer cells. B. Prostate cancer cells are placed in transwell inserts, & CM of U937 & U937-M cells is added. C. Prostate cancer cells are placed in transwell inserts with Matrigel®-coated membranes in the upper compartment, & U937-M cells are placed in the lower compartments. After 24-h incubation, the cells that had migrated through the membrane are stained. The mean optical density (OD) value is read using a microreader at 595 nm. Data are presented as mean ± SD. D. Chemokine arrays comparing the CM of PC-3 cells in monoculture & the CM of PC-3 cells co-cultured with monocyte cells. E. Prostate cancer cells are co-cultured with U937 cells for 24 h, CM is collected, & CCL2 levels are analyzed using ELISA. Adjustments of brightness, contrast, & size are applied to the whole images of western blot-based analyses without elimination of any information present in the original, including backgrounds. The mean OD value is read using a microreader at 450 nm, & data are presented as mean ± SD. All experiments are performed in triplicate, & the mean values are shown. *p < 0.05, **p < 0.01, ***p < 0.001. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28039457), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HMGB1-induced EMT in human airway epithelial cells.Human primary airway epithelial cells & BEAS-2B cells were treated with different doses of HMGB1 for mRNA analysis at different time points, & at 24 h for protein analysis. E-cadherin, ZO-1, & vimentin mRNA expression was detected by real-time quantitative PCR in (a) human primary airway epithelial cells & (c) in BEAS-2B cells after HMGB1 treatment (300 ng/mL) for 24 h. E-cadherin, ZO-1, & vimentin protein expression in (b) human primary airway epithelial cells andBEAS-2B cells (d) was assessed by western blot analysis. Data are expressed as mean ± SD (n = 3–5). *p < 0.05, **p < 0.01, ***p < 0.001, as compared with the control group. (e) TGF-beta 1 expression in cell lysate (LAP) was assessed by western blot analysis & in cell medium (active form) by ELISA. Quantification of protein expression was performed using ImageJ software. Data are expressed as mean ± S.D. (n = 4). In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots were incubated with different primary antibodies for analysis of EMT markers expression. Original blots of (b) are presented in Supplementary Fig. 3. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Expression of CSE in human aortic valves. (a) Western blot of CSE protein from AS valve (N = 12) lysates & AI valve (N = 9) lysates (left panel) were performed. Representative CSE protein levels of three AS & three AI valves lysates were shown. Relative CSE levels were assessed by densitometry analyses of band intensities of CSE western blots normalized to GAPDH (panel right). (b) Zn2+ precipitated sulfide content under alkaline conditions was measured in AI (N = 18) & AS (N = 18) valve tissues normalized to protein content of the individual samples. (c) VIC derived from different AI (N = 6) & AS (N = 9) patients were cultured in calcification medium. CSE expression was measured after the initiation of calcification. Results were normalized to GAPDH of the samples. (d) Double immunohistochemistry of CSE‐SMA & CSE‐ALP was shown with two different magnifications (×400 magnification & ×1,000 magnification). Data shown are means ± SEM of N experiments. *P < .05, significantly different as indicated Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31017307), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Demonstration of the advantages of crude fractionation over whole cell lysis & scale up of the approach. (A) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well plate & subsequently lysed directly in NP40 or RIPA lysis buffer or processed according to the protocol in Figure 2 (Sequential). An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by staining with Coomassie blue or by Western blotting & probing with antibodies to various marker proteins of specific intracellular organelles as previously. (B) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish & then transfected the next day in triplicate with 400 ng of a construct encoding V5/6×-HIS tagged ERGIC-53. Cells were extracted 48 hours later either directly in RIPA buffer, directly in NP40 buffer or according to our extraction protocol. Proteins were then partially purified from each extract using Nickel resin (SIGMA #P6611) & eluted proteins were analyzed by 10% SDS PAGE followed by staining with Coomassie blue or by Western blotting & probing with the anti V5 antibody. (C) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish & then transfected the next day in with 1600 ng of pcDNA6/V5-His (Invitrogen) or constructs encoding WT or MUT COMP. Cells were harvested 48 hours later & extracted according to our protocol. (D) HEK293 cells were plated at a density of 4 × 105 cells per well of a 12 well dish or 8.4 × 105 cells per well of a 35 mm dish & processed according to the protocol in Figure 2. An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by Western blotting & probing with antibodies to GAPDH, BiP & Lamin A. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20003239), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Effect of forskolin on the surface expression of CFTR. (A) Quantitation & representative blot of cell surface biotinylation of CFTR in HEK‐CFTR cells that were treated with forskolin (FSK; 10 μmol/L) for 1, 4, & 10 min. GAPDH was used as a control to determine if there was any contamination of biotinylated protein in the nonbiotinylated fractions (Ab: monoclonal mouse‐anti‐CFTR‐C & anti‐GAPDH; 1:1000 & 1:2000 dilution, respectively; estimated sizes: for CFTR: 165 kDa & GAPDH: 36 kDa). (B) Effect of FSK on CFTR location in live HEK‐CFTR cells. Cells grown in 35‐mm glass‐bottom dishes were stained as described in the Methods. Images were captured before & after the treatment of FSK as time series. Snapshots at 0, 1, 4, & 10 min are shown. Green: CFTR; red: WGA; & blue: Dapi. White arrows: colocalization of CFTR & WGA; green arrows: CFTR vesicles. n ≥3 for all experiments. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25263207), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HMGB1-activated AKT/GSK3 beta/ beta-catenin signaling pathways.(a) BEAS-2B cells were treated with different doses of HMGB1 for 10 min & expression of different proteins was detected by western blot. (b) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for expression of different proteins by western blot. (c) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin expression by western blot. (d) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin mRNA expression by real-time quantitative PCR. (e) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for nuclear & cytosolic beta-catenin expression by western blot. (f) BEAS-2B cells were treated with 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated with HMGB1 (300 ng/mL) for 24 h. beta-catenin expression was assessed by western blot analysis. Quantification of protein expression was performed using ImageJ software. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, as compared with the control group. #p < 0.05, as compared the HMGB1-treatment group. (A,B) (n = 4); (C–F) (n = 3). (g) Immunofluorescence staining of beta-catenin (green) was detected by fluorescence microscopy. Data are expressed as mean ± SD (n = 4). *p < 0.05, as compared with the 0-min group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots were incubated with different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HER2/HER3 & AXL expression & phosphorylation analysis.A) Representative western blots of total cell lysates of HCC827 & HCC4006 parental cell lines (P) & their derived ERL-resistant cell lines. Arrows indicate the expected molecular weight size. Total cell lysates loaded were 40 μg for AXL & pAXL analyses & 25 μg for the others. B) qPCR analysis of AXL mRNA normalized to rp-L31 mRNA & expressed relative to the levels in parental cell lines set as 1 (mean ± SD of triplicate determinations). Western blots & qPCR data are representative of those obtained respectively from 3 & 2 independent analysis. C) Dose-effect curves were calculated using CompuSyn software & plotting the entered Fa values against the entered dose values. For combination treatments, the combined drugs dose was entered. Each data point represents the mean of 3 replicates. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0143333), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Ang II positively regulates VEGF-A-mediated EC differentiation: the effect of Ang II on VEGF-A-stimulated differentiation of MSCs into ECs was examined. AT2R & VEGFR-2 mRNA expression was analyzed by RT-PCR (n = 3) (a). Sox18 protein levels were measured by Western blot analysis (n = 3) (b). Expression of EC markers was determined by FACS analysis, & a representative grid is shown (n = 5-6) ((c)–(g)). Endothelial tube formation was examined using an angiogenesis assay (n = 3) ((d)–(h)). Experiments were performed with samples taken from independent BM-MSC cultures from separate microswine. HUVECs were excluded from statistical analyses. Data are shown as mean ± SD. ∗p < 0.05 versus naïve MSCs & #p < 0.05 MSCs treated with VEGF-A versus VEGF-A plus Ang II cotreatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26106428), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - HMGB1-activated AKT/GSK3 beta/ beta-catenin signaling pathways.(a) BEAS-2B cells were treated with different doses of HMGB1 for 10 min & expression of different proteins was detected by western blot. (b) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for expression of different proteins by western blot. (c) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin expression by western blot. (d) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for beta-catenin mRNA expression by real-time quantitative PCR. (e) BEAS-2B cells were treated with HMGB1 (300 ng/mL) for different periods & analyzed for nuclear & cytosolic beta-catenin expression by western blot. (f) BEAS-2B cells were treated with 10 μM PI3K inhibitor (LY294002) & GSK-3 beta inhibitor (SB415286) for 10 min, & then treated with HMGB1 (300 ng/mL) for 24 h. beta-catenin expression was assessed by western blot analysis. Quantification of protein expression was performed using ImageJ software. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, as compared with the control group. #p < 0.05, as compared the HMGB1-treatment group. (A,B) (n = 4); (C–F) (n = 3). (g) Immunofluorescence staining of beta-catenin (green) was detected by fluorescence microscopy. Data are expressed as mean ± SD (n = 4). *p < 0.05, as compared with the 0-min group. In immunoblotting assay, gels have been run under the same experimental conditions. Then cropped blots were incubated with different primary antibodies for analysis of signaling pathway. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26739898), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Altered Rho GTPase signaling in amygdala of RICH2 KO mice. (A) Comparison between the GTPase activating protein (GAP) activity of the P2 lysates from WT & RICH2 KO mice. The GAP activity is measured in nmoles/min/mg of small G-protein. First a standard curve is made using the absorbance of KH2PO4 at different concentrations, later the absorbance of WT & RICH2 KO lysates are plotted on the graph. The GAP activity is higher for WT lysates compared to RICH2 KO lysates. (B) In the presence of the small G protein RhoA, the absorbance of WT lysates is higher than that of RICH2 KO lysate, which indicates that RICH2 is acting as a GAP for RhoA in amygdala. (C) There is no difference between the absorbance of WT & RICH2 KO lysates in the presence of RAC1 & CDC42 (D,E) Like RAC1 & CDC42, RAS p21 is not a target G protein for RICH2 in amygdala as there is no difference in the absorbance of WT & RICH2 KO lysates in the presence of RAS p21. (F) Western blot analysis of amygdala P2 lysates reveals that expression of RhoA is significantly higher in RICH2 KO lysates, in comparison with WT lysates (Unpaired t-test, p = 0.0508, n = 3). (G,H) Western blot analysis revealed no alteration in the expressions of Rac1 (G) & Cdc42 (H) between WT & RICH2 KO using amygdala P2 lysates. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/article/10.3389/fnmol.2017.00180/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Expression of Npr2 & cGKI alpha in DRGs at different developmental stages. (A,B) Western blot of membrane or cytosolic fractions of DRGs using antibodies to Npr2 or to cGKI alpha, respectively. The heavy chain of clathrin or GAPDH served as loading control. (C–E) Sections through thoracic DRGs at different stages. Localization of anti-beta Gal staining indicating Npr2-expression is demonstrated in the Npr2wt/LacZ-mouse reporter & localization of cGKI alpha by a polyclonal antibody to mouse cGKI alpha. Scale bars in (C1,D1), 50 μm; in (C2-C5,D2-D5), 100 μm; in (E) 25 μm. (F) Quantification of Npr2-positive & cGKI alpha-positive neurons in DRGs counterstained with an antibody to PGP9.5 at different stages. (G–I) A small proportion of Npr2-positive DRG neurons at P45 & P75 express trkA, parvalbumin, CGRP or IB4. Scale bars in (H,I), 20 μm. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29472841), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Bub1-KD & Bub1-T589A display increased residency in the cytosol.(a) Mitotic control (siGL2) & Bub3-depleted (siBub3) cells were fixed & stained with anti-H2A-pT120 (red), anti-Sgo1 (green) & anti-CREST (blue). (b) Quantification of the localization of H2A-pT120 & Sgo1 signals. Data represent the mean±s.e. of three independent experiments. Eighty to 300 cells were scored per condition per experiment. (c) Images & (d) quantification (normalized average pixel intensity); low (1–1.2), medium (>1.2 to ≤1.3) & high (>1.3) of 3 × MYC-GFP-Bub1 signal & localization in live cells synchronized in mitosis by a thymidine release. Data represent the mean±s.e. of 3 independent experiments, with 58–61 cells measured per condition. Significance was measured for the high group by one-way analysis of variance (ANOVA) & pairwise t-test (Holm–Sidak). (e) Scatter plot of the cytoplasm versus kinetochore GFP levels of individual cells from each of the stable cell lines. The number of cells, R2 (measure of the goodness-of-fit) & significance (one-way ANOVA) are indicated. (f) Western blottings showing levels of the 3 × MYC-GFP-Bub1 proteins in the stable cell lines in whole cell extracts (left) & in cytoplasmic extracts (right). Scale bar, 10 μM. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/ncomms9364), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Notch1, ApoER2 & Dab1 colocalize in hippocampal neurons. (A) Representative image of fluorescent immunolabeling shows that Notch1 is localized in a subset of CA3 neurons, co-expressing Dab1 & ApoER2 (magnified insert). ApoER2 is highly expressed in all CA3 neurons. (B) Fluorescent immunolabeling shows the distribution of Notch1 & Dab1 as compared to Arc/Arg3.1 in CA3 neurons. (B′) Close up of a dendrite showing colocalization of the three proteins (white arrowheads). (C) Western blot analysis on immunoprecipitated samples from whole hippocampal lysate shows that Notch1 displays a strong interaction with Dab1 whereas Notch1 & ApoER2 interaction appears weaker in the preparation (n = 4 independent experiments). (D) Immunolabeling on immunoprecipitated samples from whole cortical lysates show that Notch1 & Dab1 interact. ApoER2 is bound to both Notch1 & Dab1 (n = 3 independent experiments). (E) Co-IP from synaptosomal membrane fractions reveals a stronger interaction between Notch1 & ApoER2, which is reduced in the N1cKO (n = 3 independent pulldowns). In (C–E) GAPDH is used as a loading control for the inputs & to detect contamination in the IP samples. Scale bar in (A) is 50 μm & (B) 25 μm. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26635527), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Reduced expression of UNC5A in FancC−/− cells. a Reduced expression of Unc5A in brain cortex of FancC−/− mice compared to wild-type littermates. Western blotting was performed with the indicated antibodies. Each lane represents a different animal (WT: n = 6; FancC−/−: n = 8). b Bar graph represents ratio of Unc5A normalized to tubulin from WT (n = 6) & FancC−/− mice (n = 8). c Bar graph representation of UNC5A expression normalized to Sdha & Tbp from wild-type (WT) & FancC−/−-derived fibroblasts (n = 3). RLU: Relative light units. d–f HEK293T cells were transfected with UNC5AICD, FANCE & FANCC constructs expressing full-length FANCC (in d), FANCC1–306 (in e) or FANCC307–558 (in f). Constructs were transfected at a molecular ratio of 1:1:1 or with 10 times the molar amount of FANCE, FANCC or UNC5AICD as indicated in the figure. The total amount of transfected plasmid was equalized for all strategies with control empty vectors. Representative immunoblots performed with anti-HA (UNC5AICD), anti-FANCC, anti-FANCE or anti-GAPDH antibodies are shown. Arrows indicate appropriate protein bands. Bar graphs represent the mean fold change ± SEM of UNC5A protein levels normalized to GAPDH compared to 1:1:1 transfection controls in at least 4 separate experiments. *p < 0.05; **p < 0.005 Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30213274), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Depletion of RAD21 & other cohesin subunits leads to differentiation of ESCs.(A): qPCR analysis of esiRNA knock-down efficiency for indicated genes (48 h RNAi, n = 3, error bars denote s.d. & *, ** indicate p<0.05 & 0.01 respectively). Transfection of a non-targeting esiRNA (Luc) was used as a control. Numbers adjacent to the gene names indicate independent, non-overlapping esiRNAs transfected. (B): Western Blot analysis of esiRNA knock-down efficiency for indicated genes (48 h RNAi; esi-1 & esi-2 indicate independent, non-overlapping esiRNAs transfected). Transfection of a non-targeting esiRNA (Luc) was used as a control. GAPDH served as a protein loading control. (C): Alkaline phosphatase staining of ESCs, which had been transfected with esiRNAs targeting RAD21, SMC1a & SMC3 (72 h post RNAi) showed a strong differentiation phenotype compared to a control (Luc). Nanog depletion served as a positive control for ESC differentiation. Scale bars correspond to 100 µm. (D): Quantification of alkaline phosphatase staining (n = 3) separated into undifferentiated (green), mixed (yellow) & differentiated colonies (red). (E+F): qPCR validation of expression changes of (E) stem cell maintenance genes & (F) lineage marker genes upon knock-down of RAD21 (two independent esiRNAs) & Luc control (48 h RNAi, n = 3, error bars denote s.d. & *, **, *** indicate p<0.05, 0.01 & 0.001 respectively). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/21589869), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Application of the approach to other cell lines. (A) HT1080 cells (2 × 105) & HeLa cells (1 × 105) were plated in the wells of a 12 well plate & the digitonin concentration required for optimal extraction of the cytosol was determined as in Figure1. (B) The remainder of the protocol from Figure 2 was then applied using 100 μg/ml digitonin as the optimal cytosolic extract in each case. An aliquot of each extract was then analyzed by 4-12% SDS PAGE followed by Western blotting & probing with antibodies to GAPDH, BiP & Histone H3 (Used as an alternative nuclear marker due to weakness of the Lamin A signal in HeLa cells). (C) Samples were analyzed by 4-12% SDS PAGE followed by staining with Coomassie blue & then silver staining showing the difference in protein composition of each sample. The bracket alongside the gels marks the position of Histones in the RIPA nuclear extract & the asterisk next to the silver stained gels denotes protein bands unique to the E-RIPA extracts. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/20003239), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: GAPDH Antibody (1D4) [NB300-221] -
Western Blot: GAPDH Antibody (1D4) [NB300-221] - Role of the TRIM11-USP14 axis in cell growth & apoptosis. a Levels of endogenous TRIM11 in HCT116 cells grown at 43 °C for 0 or 90 min, or at 43 °C for 90 min, & then at 37 °C for 60 or 120 min. b, c Levels of endogenous TRIM11 (b), or Flag-TRIM11, the indicated proteasome subunits, & USP14 (c) in the SN & P fractions of HCT116 cells with & without heat shock at 43 °C for 90 min. d, g TRIM11 overexpression (d) or knockdown (g) HCT116 cells & the corresponding control cells grown under unstressed, heat shock, & heat shock plus recovery conditions were analyzed for K48 polyUb conjugates & caspase-3 activation. e, f, h Apoptosis in TRIM11 overexpression (e, f) or knockdown (h) HCT116 cells, & the corresponding control HCT116 cells, treated with & without heat shock. i, j Caspase-3 activation (i) & apoptosis (j) in HCT116 cells expressing the indicated proteins and grown under normal or heat stress conditions. k–m K48 polyUb conjugates, caspase-3 activation (k), amyloid-like fibrils (l), & apoptosis (m) in HCT116 cells expressing the indicated proteins and grown under normal & heat stress conditions. In e, f, h, j, l, & m, data represent mean ± SEM (n = 3 unless otherwise indicated). *P < 0.05; **P < 0.01; ***P < 0.001. Uncropped blots are presented in Supplementary Fig. 14 Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41467-018-03499-z), licensed under a CC-BY license. Not internally tested by Novus Biologicals.

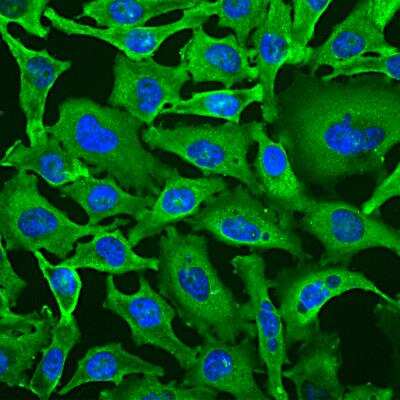
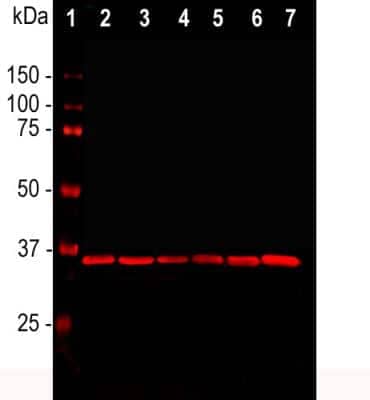
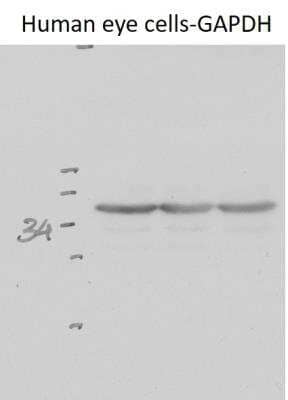
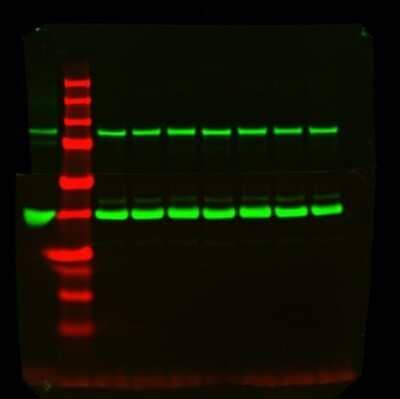
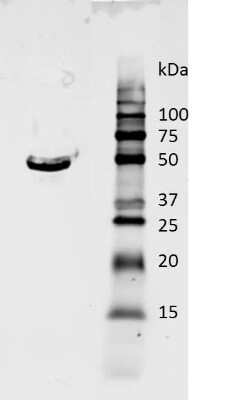
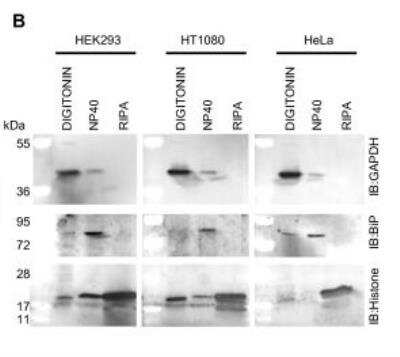
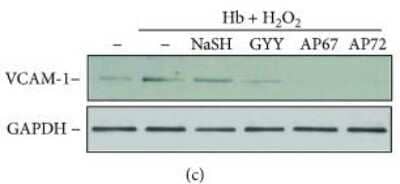
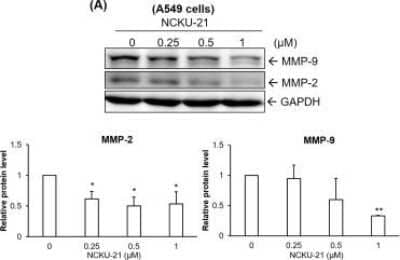
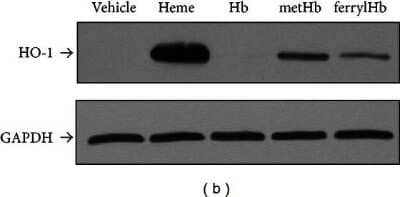
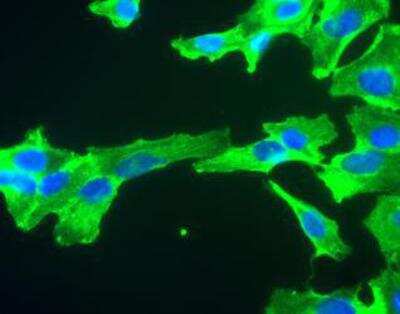
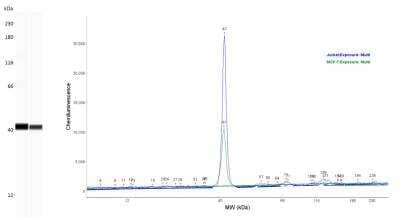
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-210202423454823.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415291920.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241535615.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241534533.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415363739.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241534330.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415395937.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415384181.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241535659.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415392547.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415384160.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415291951.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241517523.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241534355.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415392565.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415392596.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415293337.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415392580.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415293312.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415293331.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202415291947.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212354.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205117.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205139.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-3102024162182.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205155.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212340.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212365.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212335.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205133.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212318.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205159.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212323.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212333.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212358.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205146.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212377.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205193.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212390.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205124.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205199.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205176.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205180.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212364.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205196.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621843.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205178.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621820.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205157.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212387.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621830.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621236.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212342.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621235.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205166.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205160.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621847.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241621822.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241620511.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212360.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205127.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205191.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-31020241620515.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-3102024162189.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205118.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212394.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212322.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205131.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416203715.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416205163.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-3102024162187.jpg)
![Western Blot: GAPDH Antibody (1D4) [NB300-221] - GAPDH Antibody (1D4)](https://resources.bio-techne.com/images/products/nb300-221_mouse-monoclonal-gapdh-antibody-1d4-imgenex-img-5019a-310202416212324.jpg)