SARS-CoV-2 Spike RBD Antibody
R&D Systems, part of Bio-Techne | Catalog # MAB10890
Recombinant Monoclonal Antibody.

Key Product Details
Species Reactivity
SARS-CoV-2
Applications
Immunohistochemistry, Neutralization
Label
Unconjugated
Antibody Source
Recombinant Monoclonal Rabbit IgG Clone # 2820H
Product Specifications
Immunogen
Human embryonic kidney cell HEK293-derived SARS-CoV-2 Spike RBD
Ala319-Phe541
Accession # YP_009724390.1
Ala319-Phe541
Accession # YP_009724390.1
Specificity
Detects SARS-CoV2 Spike RBD in ELISA.
Clonality
Monoclonal
Host
Rabbit
Isotype
IgG
Scientific Data Images for SARS-CoV-2 Spike RBD Antibody
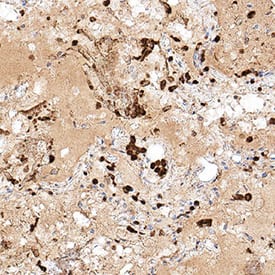
Detection of Spike RBD in SARS-CoV-2 infected human lung.
Spike RBD was detected in immersion fixed paraffin-embedded sections of SARS-CoV-2 infected human lung using Rabbit Anti-SARS-CoV-2 Spike RBD Monoclonal Antibody (Catalog # MAB10890) at 3 µg/mL for 1 hour at room temperature followed by incubation with the Anti-Rabbit IgG VisUCyte™ HRP Polymer Antibody (Catalog # VC003). Before incubation with the primary antibody, tissue was subjected to heat-induced epitope retrieval using Antigen Retrieval Reagent-Basic (Catalog # CTS013). Tissue was stained using DAB (brown) and counterstained with hematoxylin (blue). Specific staining was localized to immunoreactive profiles scattered throughout the tissue. View our protocol for IHC Staining with VisUCyte HRP Polymer Detection Reagents.Applications for SARS-CoV-2 Spike RBD Antibody
Application
Recommended Usage
Immunohistochemistry
1-3 µg/mL
Sample: Immersion fixed paraffin-embedded sections of SARS-CoV-2 infected human lung
Sample: Immersion fixed paraffin-embedded sections of SARS-CoV-2 infected human lung
Neutralization
In a functional ELISA binding assay, 40.0-480 ng/mL of this antibody will block 50% of the binding of 50 ng/mL of Recombinant Human ACE 2 (Catalog # 933-ZN) when Recombinant SARS-CoV-2 Spike RBD His tag (HEK293 Expressed)(Catalog # 10500-CV) is immobilized at 0.2 ug/mL (100 µL/well). At 5 ug/mL, this antibody will block >90% of the binding.
Formulation, Preparation, and Storage
Purification
Protein A or G purified from hybridoma culture supernatant
Reconstitution
Reconstitute at 0.5 mg/mL in sterile PBS. For liquid material, refer to CoA for concentration.
Formulation
Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. *Small pack size (SP) is supplied either lyophilized or as a 0.2 µm filtered solution in PBS.
Shipping
Lyophilized product is shipped at ambient temperature. Liquid small pack size (-SP) is shipped with polar packs. Upon receipt, store immediately at the temperature recommended below.
Stability & Storage
Use a manual defrost freezer and avoid repeated freeze-thaw cycles.
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: Spike RBD
References
- Wu, F. et al. (2020) Nature 579:265.
- Tortorici, M.A. and D. Veesler (2019). Adv. Virus Res. 105:93.
- Bosch, B.J. et al. (2003) J. Virol. 77:8801.
- Belouzard, S. et al. (2009) Proc. Natl. Acad. Sci. 106:5871.
- Millet, J.K. and G. R. Whittaker (2015) Virus Res. 202:120.
- Yuan, Y. et al. (2017) Nat. Commun. 8:15092.
- Walls, A.C. et al. (2010) Cell 180:281.
- Jiang, S. et al. (2020) Trends. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Ortega, J.T. et al. (2020) EXCLI J. 19:410.
- Wrapp, D. et al. (2020) Science 367:1260.
- Tai, W. et al. (2020) Cell. Mol. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Okba, N. M. A. et al. (2020). Emerg. Infect. Dis. https://doi.org/10.3201/eid2607.200841.
- Wang, X. et al. (2020) https://doi.org/10.1038/s41423-020-0424-9.
- Wang, K. et al. (2020) bioRxiv https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1.
Long Name
Spike Receptor Binding Domain
Gene Symbol
S
UniProt
Additional Spike RBD Products
Product Documents for SARS-CoV-2 Spike RBD Antibody
Product Specific Notices for SARS-CoV-2 Spike RBD Antibody
For research use only
Loading...
Loading...
Loading...
Loading...