Proper laboratory preparation, sample preparation, assay running and RNA quality controls, and understanding data interpretation and assay optimization can ensure a smooth path to results when using RNAscope™ assays.
Materials Checklists and User Manuals
Our assays often require a combination of common laboratory materials, materials provided by Bio-Techne, materials provided by other vendors, and instrumentation or equipment. You can find these items listed in the user manual specific to the assay you are running, or in many cases, a pre-run checklist is available to easily review required materials.
Please Note: For ordering checklists and user manual, please go to the documents section on each individual product page to download.
A. 样本制备
正确制备样本为实现使用 RNAscope ISH 技术时有可能的高水平灵敏度和特异性奠定基础。ACD 原位杂交试剂盒(RNAscope、BaseScope™、miRNAscope™ 和 DNAscope™)可配合福尔马林固定、石蜡包埋 (FFPE) 的样本、新鲜冷冻 (FF) 或固定冷冻的组织以及贴壁培养和非贴壁培养的细胞使用。
ACD 网站上可提供制备上述每种样本类型用于各分析法类别(显色、荧光、多重分析、自动化)的最佳实践指南。“产品文档”的“支持”部分中存有刊载针对每种分析法详细建议样本制备过程的产品手册和相关技术说明。“支持解决方案”的“故障排除指南和常见问题解答”部分也刊载大量关于样本制备的信息。
User's Guide Webinar
A New User's Guide to Getting Started with RNAscope ISH Technology
RNAscope ISH Technology is the gold standard for in situ hybridization (ISH), providing spatial visualization of gene expression with unparalleled sensitivity, specificity, and reliability.
This webinar will provide a comprehensive overview of our RNAscope ISH assays to help new RNAscope users set up and get started.
We discuss the workflows for each manual assay, including required materials and reagents, tips for sample preparation and pre-treatment optimization, and guidelines for image acquisition and data analysis. We will also discuss recommendations for troubleshooting and optimization.
Running RNAscope assays with the control slides, and testing your samples with the positive and negative control probes is recommended. Staining control slides will verify the assay workflow, and testing the control probes on the test samples will verify the quality of the RNA in test samples, as well as help to assess the pretreatment conditions. The housekeeping gene ppib (Cyclophilin B), often used as a reference gene for RT-PCR, is most frequently used as a positive control. The bacterial dapB gene is used as a negative control.
Search Control Probes
Find species-specific positive control probes and negative control probes for your assays.
Pretreatment
Following appropriate sample preparation, sample pretreatment provides sufficient access for ISH target probes to hybridize to the target of interest. Our sample pretreatment reagents and guidance are designed to permeabilize the cell membrane, to partially relieve crosslinking of nucleic acid to proteins that occurs during sample fixation, and to block endogenous peroxidase activity (if needed).
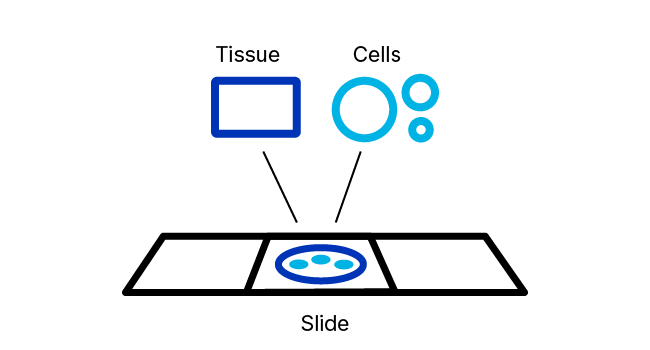
RNAscope ISH Pretreatment Reagents
RNAscope Pretreatment reagents provide the right tools for these important jobs. The proprietary formulations of these reagents have been optimized to provide enhanced access of in situ hybridization probes to nucleic acid targets across a wide variety of tissue sample types including FFPE, fresh‑frozen (FF), and fixed-frozen tissue as well as tissue microarray (TMA) and cultured adherent and non-adherent cells.
RNAscope Target Retrieval: A buffer system used along with heating of the samples to partially reverse the cross-linking that occurs from tissue fixation, including perfused tissue. This reagent is not required for fresh (unfixed) frozen tissue.
RNAscope Hydrogen Peroxide Reagent: For blocking endogenous peroxidase activity.
RNAscope Proteases: Reagent used to permeabilize cell membranes as well as to unmask RNA or DNA targets by degrading bound proteins. Different sample preparation methods may require reagents with different proteolytic activity. We offer several types of protease including RNAscope Protease Plus, Protease III, and Protease IV.
Pretreat Pro/AMP Pro: These reagents, in combination with retrieval and blocking where needed, provide a protease-free pretreatment workflow to streamline co-detection and multiomic workflows.
Probe Hybridization
After pretreatment, samples undergo an optimized probe hybridization step. For duplex and multiplex assays, probes are pooled prior to this hybridization step.
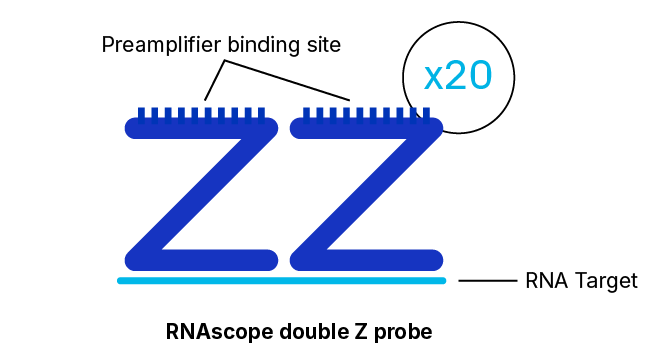
RNAscope Probe Hybridization Technology
RNAscope, BaseScope, and DNAscope assays employ a probe hybridization strategy that involves pairs of probes, each of which contains 18 to 25 bases complementary to a section of the target RNA adjacent to that of the other pair member. The overall system is designed such that both probes in each pair must hybridize independently to their respective target sequences for subsequent signal amplification to occur.
High Specificity, Excellent Signal-to-Noise Ratio
This pairing requirement not only has the effect of greatly increasing the signal-to-noise ratio but also ensures high specificity stemming from the extremely low probability of two different probes binding adjacent non-specific sequences. Furthermore, our probe design algorithm is validated to select for optimal hybridization within the recommended assay conditions and minimal cross-hybridization to off-target sequences.
Single RNA Molecule Detection Achieved with Degraded Samples or Less Accessible Targets
Although detection of a single RNA target molecule with an RNAscope assay can be accomplished by the binding of just three pairs of probes for the target, there are 20 separate pairs of probes specific to the target in the assay. This high level of redundancy means that even partially degraded or incompletely unmasked RNA targets may still be detectable at single-molecule resolution. Another feature that contributes to the robustness of RNAscope assays in this respect are the relatively short target regions required, with just 36 to 50 bases of combined target sequence being sufficient for the hybridization of a probe pair.
Amplification and Detection
The final steps in the workflow serve to the generate a highly amplified, readily detectable signal indicating the presence (and location) of the target molecule. The unique three-component design of RNAscope, BaseScope, and DNAscope hybridization probes provides the basis for sensitivity capable of detecting even a single RNA or DNA molecule.
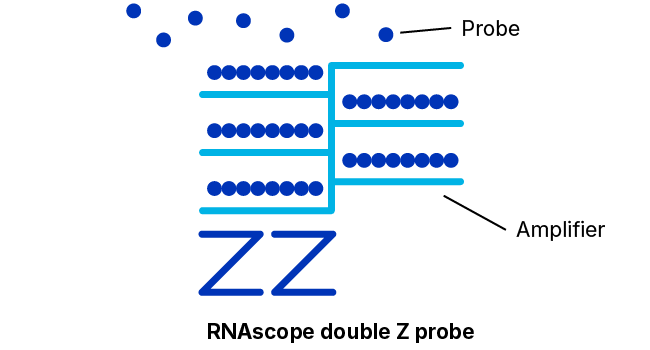
Adjacent Z-Probe Pairs Form a Binding Site for the Amplification Apparatus
The 18- to 25-base “lower” portion of each hybridization probe of the pair just discussed that binds specifically and adjacently to the target molecule is connected via a spacer sequence to a 14-base “upper” portion. The spacer sequences cause each probe construct to assume a “Z” shape wherein the two upper portions of a bound pair will also lie adjacently in a line. This upper structure forms a 28-base binding site for a so-called pre-amplifier molecule.
As additional insurance against signals arising from non-specific binding, the pre-amplifier requires both upper portions, i.e., the entire 28-base sequence, to be presented contiguously to bind.
Many Amplifiers Bind Each Pre-Amplifier; Each Amplifier Binds Many Labeled Probes
Once bound to the upper portion of the probe pair, the pre-amplifier provides a kind of scaffolding that bears numerous binding sites for “amplifier” molecules, each of which contains a similar number of binding sites for labeled probes that contain either a chromogenic enzyme or a fluorescent molecule.
The multiplicity of labeled probes arising from the binding of a single molecule is what enables the dramatic levels of signal amplification characteristic of RNAscope assays.
Detection by Microscopy
Detection is accomplished via fluorescent or brightfield microscopic analysis, depending on the type of assay selected. Each punctate dot signal represents a single target molecule. The system can also be automated on partner platforms (Leica Biosystems and Roche Tissue Diagnostics). Single-molecule signals can be quantified on a cell-by-cell basis by manual counting or automated using commercial image processing software packages. We provide technical notes for the open-source image processing programs QuPath, ImageJ, and CellProfiler.
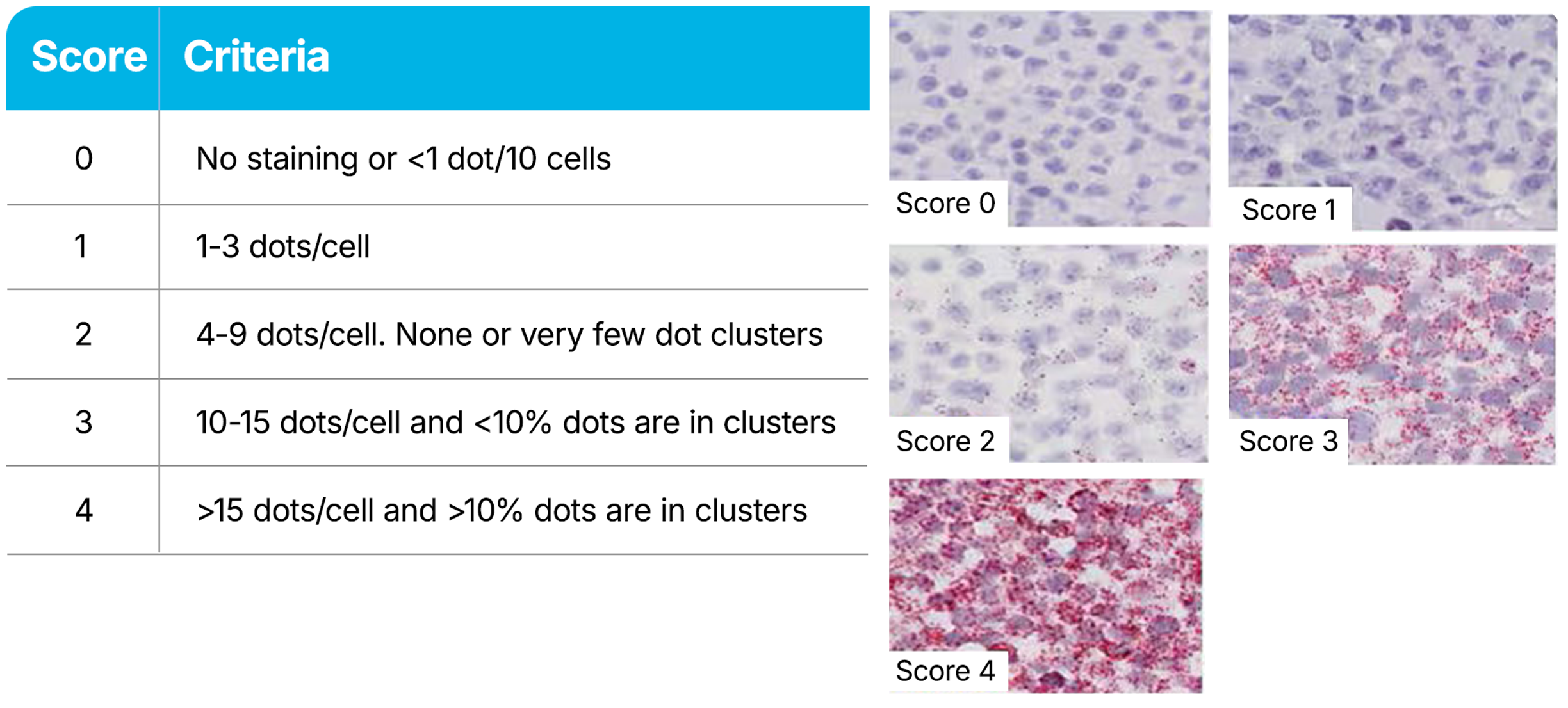
RNAscope Assays use a semi-quantitative scoring guideline to evaluate the staining results. When interpreting RNAscope staining, we recommend scoring the number of dots per cell rather than the signal intensity. The number of dots correlates to the number of RNA copies, whereas dot intensity reflects the number of probe pairs bound to each molecule. Compare the expression of your target gene with both negative (dapB) and positive controls (PPIB, UBC, or POLR2A). Successful staining should have a PPIB/POLR2A score ≥2 or a UBC score ≥3. The negative control (dapB) score should be <1.
The antigen retrieval conditions may require optimization. These conditions depend on the tissue type and how the sample was prepared. If the sample preparation differs from our protocol, we recommend starting with the pretreatment recommendations provided in the user documentation. If sample preparation details are unknown or vary from our protocol, simple optimization steps can help you obtain optimal results with the RNAscope assay.
Resources
更多了解
以下出版物详述了 RNAscope 技术背后的核心原则:
Wang F, Flanagan J, Su N, 等人 RNAscope: A Novel In Situ RNA Analysis Platform for Formalin-Fixed Paraffin-Embedded Tissues. J of Mol Diagnostics. 2012 Jan; 14(1):22-29.

