By Rosa Moreno, PhD.
Detecting HIF alpha and beyond: Best controls for hypoxia Western blot analysis
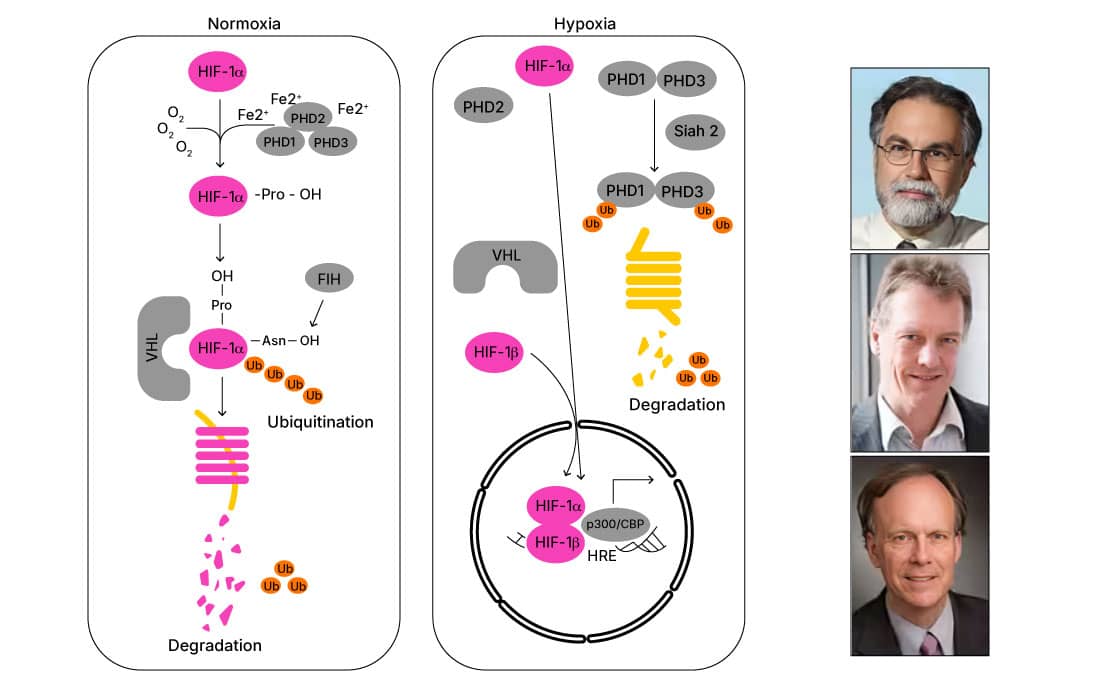
Physiological low levels of oxygen induce hypoxic events across biological systems. This hypoxic state activates hypoxia inducible factors (HIFs) to regulate transcription by binding to the hypoxia response element (HRE) region of downstream target genes. While these responses may be physiologically normal in day to day life, chronic activation of HIFs has been associated with disease states including ischemia and cancer.1 Here we discuss the regulation of HIFs under both normoxic and hypoxic events, demonstrating the importance of the use of experimental controls when assessing the effects of hypoxia.
Cellular oxygen sensing and adaptation: A Nobel Prized mechanism
HIF-1 alpha (HIF-1a) and HIF-2 alpha (HIF-2a) are key regulators of the cellular responses to low oxygenation and are often referred to as "master regulators" of hypoxic responses. HIF-1a was first identified and isolated by Dr Gregg L. Semenza in the early 1990s while studying the regulation of erythropoietin (EPO) expression by oxygen.2 The oxygen-dependent modulating factor was then recognized to consist of DNA binding HIF-1a and ARNT (aryl hydrocarbon receptor nuclear translocator) or HIF-1 beta subunits. Studies by Dr Semensaz's and Dr Peter J. Ratcliffe's groups further helped to uncover the ubiquitous nature of the identified oxygen sensing mechanism.
Nobel Prize in Physiology or Medicine 2019 recipients. From top:
Gregg L. Semenza, Peter J. Ratcliffe and William G. Kaelin
Under normoxic conditions, HIF-1a is rapidly (~5-minute half-life) hydroxylated by prolyl hydroxylases (PHD enzymes), which together with von Hippel-Lindau protein (pVHL), target HIF-1a for proteasomal degradation. However, during hypoxic events, HIF alphas translocate to the nucleus where they form a stable heterodimer with HIF beta. By interacting with p300/CBP co-activators, this complex regulates the expression of HRE-containing target genes.1 The mechanisms regulating HIF-1a were uncovered by Dr William G. Kaelin's and Dr Ratcliffe's work in the early 2000s. Their work demonstrated the key hydroxylation events allowing pVHL’s association with HIF alpha and its consequent proteasomal targeting, and thus explained the molecular basis of cellular oxygen sensing.3, 4
Application Focus: Preserving HIF alpha during sample preparation
Given HIF alphas are targeted for ubiquitination in the presence of oxygen, they are rapidly degraded when exposed to room air. Thus, care must be taken during sample preparation when assessing HIF expression by Western blot. However, to mitigate the degradation process, HIF proteins can be stabilized during sample preparation using protease inhibitors or CoCl2 in buffers for cell homogenization.5 Mechanistically, CoCl2 prevents hydroxylation of HIF alpha by inhibiting the activity of PHD enzymes. Additionally, CoCl2 directly binds to HIF-2a, inhibiting its interaction with pVHL and thus stabilizing HIF levels.6,7 It has been demonstrated that inclusion of CoCl2 alone during sample preparation (i.e., without protease inhibitors) is sufficient to preserve HIF levels, providing an effective yet inexpensive approach for HIF stabilization.5,7
In the absence of hypoxia mimetic agents, preparing cell lysates to analyze HIF alpha levels requires swift sample handling. For cells in culture, media is quickly removed, cells are washed with cold PBS on ice and scraped directly in lysis buffer to prevent HIF degradation.
Best controls for HIF antibody validation
Treatment or incubation of cells with CoCl2 is an approach often used to mimic hypoxia, which provides the benefit of using normoxic-conditions in experimental setups. As a biological validation strategy, cellular lysates from cell lines (e.g., HeLa, HepG2 and Cos7 cells) exposed to low oxygenation or CoCl2 are commonly used in order to verify the specificity and sensitivity of antibodies to HIF-1a or HIF-2a in Western blot applications. Inclusion of these controls in your Western blot analysis ultimately facilitates troubleshooting unexpected results such as the absence of a HIF alpha band in experimental samples or the presence of unexpected bands at a lower molecular weight than expected which may be due to degradation (e.g., ~60kDa band rather than the expected ~120kDa band). Moreover, control lysates provide a reliable setting to evaluate the induction of other proteins whose expression is regulated by hypoxia and/or HIF alphas.
| Control Lysates | Preparation Notes |
|---|---|
|
Cell lysates from HeLa cells incubated under normoxic or hypoxic conditions (NBP2-36452) were analyzed for HIF-1a expression by Western blot with HIF-1a antibody (NB100-134). Tubulin was used as a loading control. |
Whole cell lysate of HeLa cells grown under normoxic-21% or hypoxic conditions- 2% oxygen for 4 hours |
|
HeLa (CoCl2 treated/untreated pair)
Cell lysates from HeLa cells untreated or treated with CoCl2 (NBP2-36450) were analyzed for HIF-1a expression by Western blot with HIF-1a antibody (NB100-134). Tubulin was used as a loading control. |
Whole cell lysate of HeLa cells cultured under standard conditions or treated with 100 μM CoCl2 for 4 hours |
|
Cell lysates from HepG2 cells incubated under normoxic or hypoxic conditions (NBP2-36453) were analyzed for HIF-2a expression by Western blot with HIF-2a antibody (NB100-122). Tubulin was used as a loading control. |
Whole cell lysate of HepG2 cells grown under normoxic-21% or hypoxic conditions- 2% oxygen for 4 hours |
|
HepG2 (CoCl2 treated/untreated pair)
Cell lysates from HepG2 cells untreated or treated with CoCl2 (NBP2-36451) were analyzed for HIF-2a expression by Western blot with HIF-2a antibody (NB100-122). Tubulin was used as a loading control. |
Whole cell lysate of HepG2 cells cultured under standard conditions or treated with 100 μM CoCl2 for 4 hours |
|
Cos7 (CoCl2 treated/untreated pair)
Cell lysates from COS-7 cells untreated or treated with CoCl2 (NB800-PC26) were analyzed for HIF-1a expression by Western blot with HIF-1a antibody (NB100-105). |
Nuclear lysate of COS-7 cells grown under standard conditions or treated for 16 hours with 150 μM CoCl2 |
Caveats to the use of CoCl2 to mimic hypoxia
To understand cellular adaptations to hypoxia it's necessary to evaluate the expression of downstream protein targets that are regulated by HIFs. While CoCl2 treatment stabilizes HIFs to mimic hypoxic states, this chemical induction may not fully replicate the expression patterns induced by low oxygen conditions or true hypoxia. Differential gene transactivation has been observed by several groups, indicating that CoCl2 treatment engages signaling pathways beyond those responsive to HIF and hypoxia. In other cases, CoCl2 treatment fails to regulate genes whose expression is modulated by low oxygenation.7
Beyond detecting HIF-1a: Detection of HIF regulated targets
HIF alpha's stabilization under hypoxic conditions results in transactivation of HIF alpha target genes which are implicated in various physiologically relevant processes.1 Among the wide array of genes containing HREs and regulated by hypoxic conditions, Carbonic Anhydrase 9 (CAIX) serves as the predominant marker for hypoxia in part due to its high responsiveness to low oxygen conditions and stable expression.8 Immunohistochemical evaluation of CAIX in tumor tissue has been used as a reporter for hypoxic conditions and a meta-analysis of multiple clinical studies (over 100) supports the prognostic value of evaluating CAIX expression.9
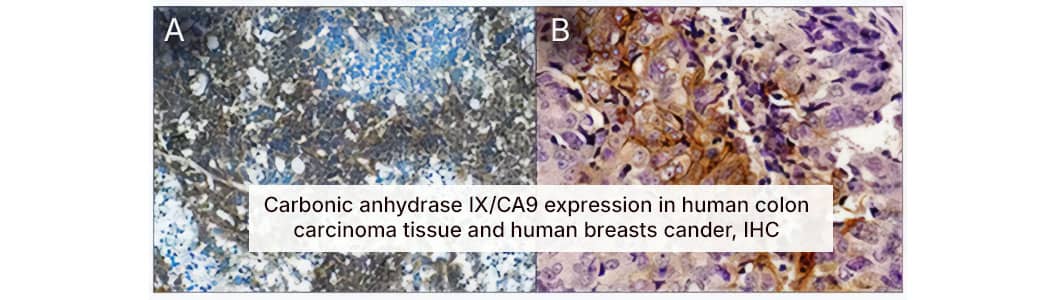
Cytoplasmic expression of Carbonic anhydrase IX/CA9 in human carcinoma tissues shows enhanced signal around hypoxic tumor cores. A. Immunohistochemical detection of Carbonic Anhydrase IX/CA9 Antibody [NB100-417] in human colon carcinoma tissue, xenografted in mice. B. Immunohistochemical detection of Carbonic Anhydrase IX/CA9 Antibody [NB100-417] in human breast cancer tissue. For detection of bound Carbonic Anhydrase IX/CA9 a secondary antibody labeled with HRP and DAB reagent were used. Cell nuclei were counterstained with hematoxylin.
From the seminal studies by Gregg L. Semenza, Peter J. Ratcliffe and William G. Kaelin, which uncovered the mechanisms underscoring cellular oxygen sensing, much remains to be learned about the connections between HIFs and downstream regulated genes in disease. However, understanding the molecular programs regulated by HIFs presents challenges at the experimental level. Oxygen-controlled conditions which mimic the physiological levels for cells and tissues under normal and diseased states may be challenging to replicate in most laboratories. Hypoxia mimetics represent an accessible alternative, although these agents may not fully replicate the effects of true hypoxia. The use of key controls in hypoxia research allows confirmation of sample integrity and may help to corroborate true hypoxic responses, thus streamlining experimentation and strengthening conclusions.
Rosa Moreno, PhD.
Product Marketing Specialist at Novus Biologicals LLC
-
Masoud, G.N. & Li, W. (2015) HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharmaceutica Sinica B
-
Semenza, G.L. & Wang, G.L. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Molecular and Cellular Biology.
-
Ivan, M. et al. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science.
-
Jaakkola, P. et al. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O 2 -regulated prolyl hydroxylation. Science.
-
Srinivasan, S. & Dunn, J.F. (2011) Stabilization of hypoxia-inducible factor-1α in buffer containing cobalt chloride for Western blot analysis. Analytical Biochemistry.
-
Yuan, Y. et al. (2003) Cobalt inhibits the interaction between hypoxia-inducible factor-α and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-α. Journal of Biological Chemistry.
-
Muñoz-Sánchez, J. et al. (2019) The use of cobalt chloride as a chemical hypoxia model. Journal of Applied Toxicology.
-
Potter, C. & Harris, A.L. (2004) Hypoxia inducible carbonic anhydrase IX, marker of tumor hypoxia, survival pathway and therapy target. Cell Cycle. PMID: 14712082.
-
Van Kuijk, S.J.A. et al. (2016) Prognostic significance of carbonic anhydrase IX expression in cancer patients: A meta-analysis. Frontiers in Oncology.