Recombinant SARS-CoV-2 P.1 Spike S1 His-tag Protein, CF
R&D Systems, part of Bio-Techne | Catalog # 11138-CV

Key Product Details
Product Specifications
Source
Human embryonic kidney cell, HEK293-derived sars-cov-2 Spike S1 Subunit protein
Val16-Pro681 (Leu18Phe, Thr20Asn, Pro26Ser, Asp138Tyr, Arg190Ser, Lys417Thr, Glu484Lys, Asn501Tyr, Asp614Gly, His655Tyr) with a C-terminal 6-His tag
Val16-Pro681 (Leu18Phe, Thr20Asn, Pro26Ser, Asp138Tyr, Arg190Ser, Lys417Thr, Glu484Lys, Asn501Tyr, Asp614Gly, His655Tyr) with a C-terminal 6-His tag
Purity
>95%, by SDS-PAGE visualized with Silver Staining and quantitative densitometry by Coomassie® Blue Staining.
Endotoxin Level
<0.10 EU per 1 μg of the protein by the LAL method.
N-terminal Sequence Analysis
Val16
Predicted Molecular Mass
75 kDa
SDS-PAGE
100-120 kDa, under reducing conditions.
Activity
Measured by its binding ability in a functional ELISA with Recombinant
Human ACE-2 Fc Chimera
(Catalog #
10544-ZN).
Scientific Data Images for Recombinant SARS-CoV-2 P.1 Spike S1 His-tag Protein, CF
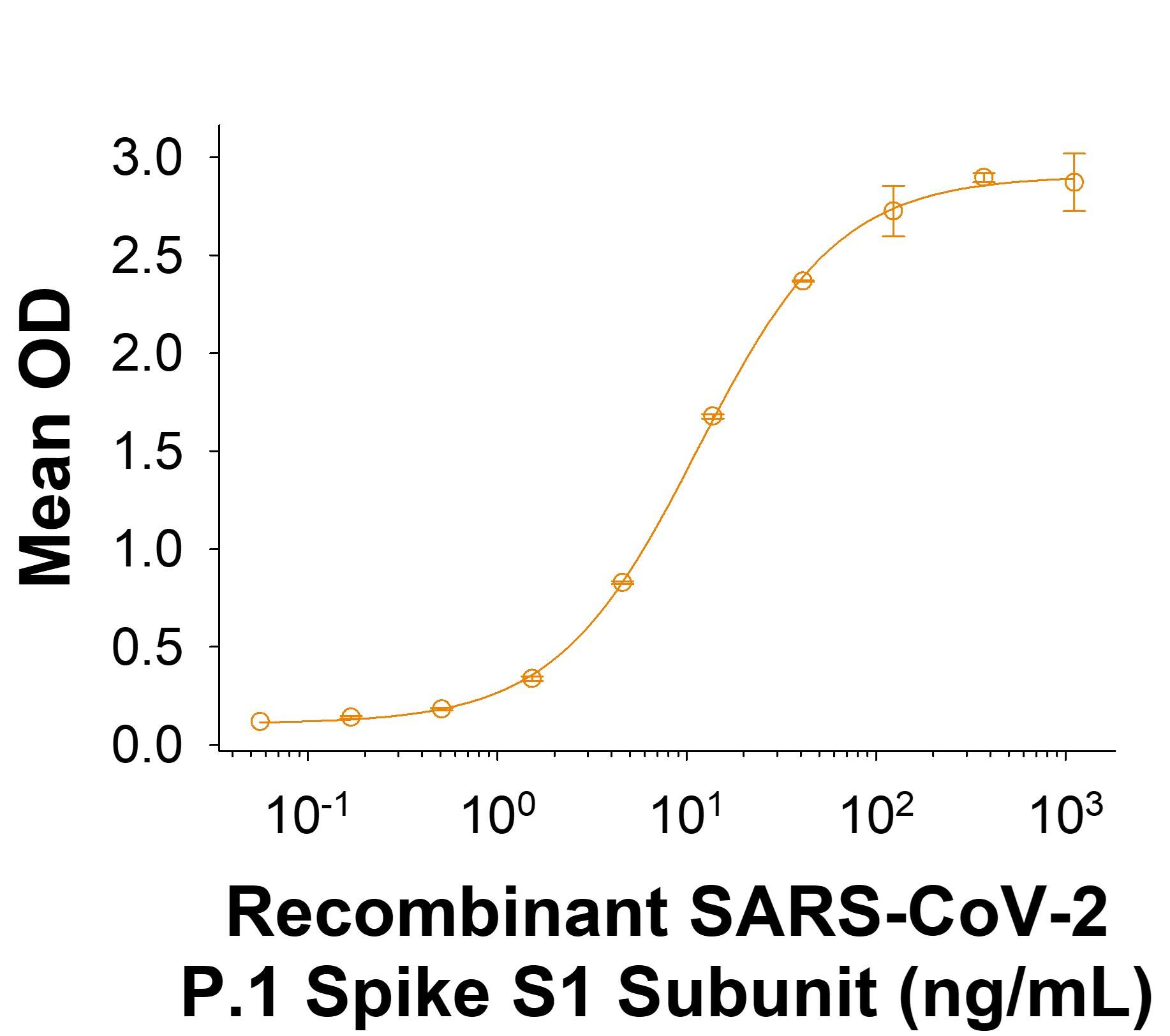
Recombinant SARS-CoV-2 P.1 Spike S1 Subunit His-tag Protein Binding Activity.
Recombinant SARS-CoV-2 P.1 Spike S1 Subunit His-tag Protein (Catalog # 11138-CV) binds Recombinant Human ACE-2 Fc Chimera (10544-ZN) in a functional ELISA.Recombinant SARS-CoV-2 P.1 Spike S1 Subunit His-tag Protein SDS-PAGE.
2 μg/lane of Recombinant SARS-CoV-2 P.1 Spike S1 Subunit His-tag Protein (Catalog # 11138-CV) was resolved with SDS-PAGE under reducing (R) and non-reducing (NR) conditions and visualized by Coomassie® Blue staining, showing bands at 100 - 120 kDa.Formulation, Preparation and Storage
11138-CV
| Formulation | Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. |
| Reconstitution | Reconstitute at 200 μg/mL in PBS. |
| Shipping | The product is shipped at ambient temperature. Upon receipt, store it immediately at the temperature recommended below. |
| Stability & Storage | Use a manual defrost freezer and avoid repeated freeze-thaw cycles.
|
Background: Spike S1 Subunit
References
- Wu, F. et al. (2020) Nature 579:265.
- Tortorici, M.A. and D. Veesler (2019) Adv. Virus Res. 105:93.
- Bosch, B.J. et al. (2003) J. Virol. 77:8801.
- Belouzard, S. et al. (2009) Proc. Natl. Acad. Sci. 106:5871.
- Millet, J.K. and G.R. Whittaker (2015) Virus Res. 202:120.
- Li, W. et al. (2003) Nature 426:450.
- Wong, S.K. et al. (2004) J. Biol. Chem. 279:3197.
- Jiang, S. et al. (2020) Trends. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Ortega, J.T. et al. (2020) EXCLI J. 19:410.
- Wrapp, D. et al. (2020) Science 367:1260.
- Tai, W. et al. (2020) Cell. Mol. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Okba, N.M.A. et al. (2020) Emerg. Infect. Dis. https://doi.org/10.3201/eid2607.200841.
- Wang, X. et al. (2020) https://doi.org/10.1038/s41423-020-0424-9.
- Wang, K. et al. (2020) bioRxiv https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1.
- Imai, M. et al. (2021) PNAS 118:e2106535118.
- Zahradník, J. et al. (2021) bioRxiv https://doi.org/10.1101/2021.01.06.425392.
- Zhang, L. et al. (2020) Nat Commun. 11:6013.
- Wang, W.B. et al. (2021) bioRxiv https://doi.org/10.1101/2021.02.17.431566.
- Nonaka, C.K.V. et al. (2021) Emerg Infect Dis. https://doi.org/10.3201/eid2705.210191.
Long Name
Spike Protein, S1 Subunit
Alternate Names
SARS-CoV-2
UniProt
Additional Spike S1 Subunit Products
Product Documents for Recombinant SARS-CoV-2 P.1 Spike S1 His-tag Protein, CF
Product Specific Notices for Recombinant SARS-CoV-2 P.1 Spike S1 His-tag Protein, CF
For research use only
Loading...
Loading...
Loading...
Loading...