Feature Products for CRISPR-based Gene Editing
CRISPR Cas9 Antibody Pack
CRISPR Cas9 Antibody Pack
Detect CRISPR-Cas proteins using our antibodies, validated for use in a range of applications.
Single Cell Isolation
Single Cell Isolation
With Bio-Techne's single cell dispensers, Hana™ and Pala™, fluorescently-labeled CRISPR gene-edited cells can be identified and rapidly and gently isolated.
Lipid Nanoparticles
Lipid Nanoparticles
We offer high quality lipids enabling you to make lipid nanoparticles for delivery of gene editing tools such as CRISPR-Cas9.
CRISPR-Cas9 Gene Editing Workflow
i. Gene Delivery
There are a variety of methods for delivery of CRISPR DNA, RNA, or RNP into cells, including lipid nanoparticles, viral and lentiviral vectors, and electroporation.
Bio-Techne offers a range of reagents for use in CRISPR delivery:
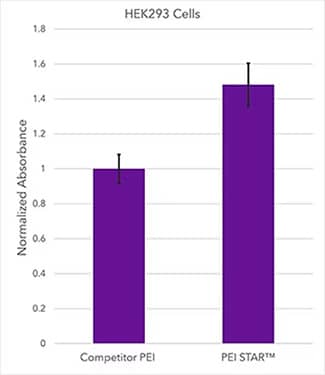
Absorbance readout of HEK293 cell suspensions transfected with a CMV-SEAP plasmid using either PEI STAR™ or competitor PEI. PEI STAR™ transfection reagent (Catalog # 7854) performance data comparison (HEK293): HEK 293 - 20 mL cultures containing HEK293 suspensions were transfected with a CMV-SEAP plasmid at optimized PEI/DNA ratios using either PEI STAR™ (3:1) or leading competitor PEI. SEAP expression levels were quantified 5 days post-transfection using phosphatase reporter dye and UV/Vis absorbance.
Efficiency of Gene Transfer
CRISPR-Cas9 antibodies can help verify the success of gene transfer. Comparing the expression of Cas9 in transfected versus mock transfected cells is important to assess the efficiency of transfection. With the help of CRISPR-Cas9 antibodies, one can use Western blot and/or immunostaining to verify transfection efficiency.
CRISPR-mediated gene editing is typically an inefficient process; the efficiency can be improved using small molecule CRISPR Enhancers.
ii. Detection of CRISPR-Cas9
Detection of CRISPR-Cas9 involves both expression analysis as well as determination of cellular localization. Analysis of these factors can be achieved through use of CRISPR-Cas9 antibodies.
CRISPR-Cas9 Expression
The levels as well as the duration of Cas9 expression are highly critical for CRISPR-Cas9 based genome editing. In stable clones, high expression levels of Cas9 may result in non-specific activity. This may be controlled through isolating multiple clones and screening them for Cas9 expression levels through Western blot analysis. For transient transfections, a chronic or prolonged expression of Cas9 may lead to the development of more off-target mutations. The transient nature of the expression of CRISPR-Cas9 may be checked through Western blot analysis of the transfected samples collected at various time points.
Subcellular Localization
CRISPR-Cas9 must translocate to the nuclei of transfected cells for executing its effects on DNA. Nuclear localization of Cas9 may be verified through ICC/IF or IHC staining with CRISPR-Cas9 antibodies.
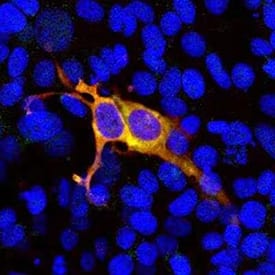
Expression of CRISPR-Cas9 in HEK293 Cells. HEK293 cells were transfected with a construct including the N-terminal 608 amino acids of S. pyogenes CRISPR-Cas9 fused to GFP and stained with Rabbit anti-CRISPR-Cas9 Polyclonal Antibody (red; Catalog # NBP3-05548). Transfected cells express the green fusion protein and bind the antibody in red, producing a yellow signal. Nuclear DNA in transfected and non-transfected cells is revealed with the blue DNA stain DAPI.
iii. Confirmation of Edits
Confirming successful editing within target cells is an important step in the CRISPR-Cas9 gene editing workflow is . Edits can be confirmed at the mRNA level using reverse transcriptase polymerase chain reaction (RT-PCR) or at the gene level by utilizing knockout (KO)-validated antibodies. Learn more about CRISPR/Cas9 Knockout Lysates
Knockout Validated Antibodies by Application
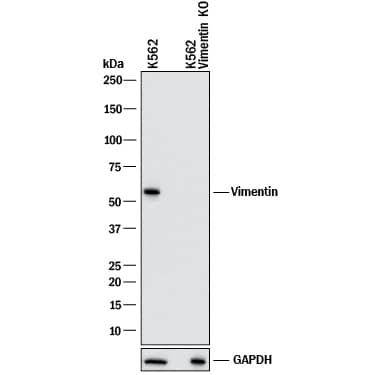
Western blot analysis of Vimentin expression in K562 parental cell line and Vimentin knockout (KO) K562 cell line. Western blot analysis showing lysates of K562 parental cell line and Vimentin knockout (KO) K562 cell line loaded at 0.2 mg/mL. The membrane was probed with Goat Anti-Human/Mouse/Rat Vimentin Antigen Affinity-purified Polyclonal Antibody (Catalog # AF2105) followed by HRP-conjugated Anti-Goat IgG Secondary Antibody (Catalog # HAF017). A specific band was detected for Vimentin at approximately 55 kDa (as indicated) in the parental K562 cell line, but is not detectable in knockout K562 cell line. GAPDH (AF5718) is shown as a loading control.
Related Products
Milo Single Cell Westerns
Single-Cell Westerns with Milo enable analysis of CRISPR-based gene editing efficiency and expression uniformity with fewer cells.
RNAscopeTM ISH Assays
RNAscopeTM ISH Assays
Advanced Cell Diagnostics, a Bio-Techne brand, offers RNAScope ISH Technology allowing for visualization and spatial detection of RNA at the single cell level, including Cas9 probes available for CRISPR-Cas9 detection.
Non-Viral Gene Engineering
Non-Viral Gene Engineering
Realize the potential of gene engineering by using TcBuster™ to deliver your genes of interest. TcBuster is a non-viral transposon system that enables stable gene transfer for the rapid generation of transgenic cells.
Related Resources
Western Blot eHandbook
Western Blot eHandbook
Our step-by-step guide to western blotting is aimed at helping you understand the principles behind this common technique and learn how to achieve reliable and reproducible results.
Single-Cell Westerns for Exploring PKC Signaling and CRISPR Efficiency
In this Webinar, Dr Noemi Kedei and Dr Kelly Gardner explain how Milo, Bio-Techne's automated single-cell western (scWestern) platform is being used to explore heterogeneity in Protein Kinase C (PKC) signaling and CRISPR knockdown efficiency.
Accelerate the Development of CRISPR-edited Cell Line Models
Learn how the Bio-Techne's Single Cell Dispenser can be used in streamlining and accelerating the development of CRISPR-edited cell line models in the whitepaper.
Background Information
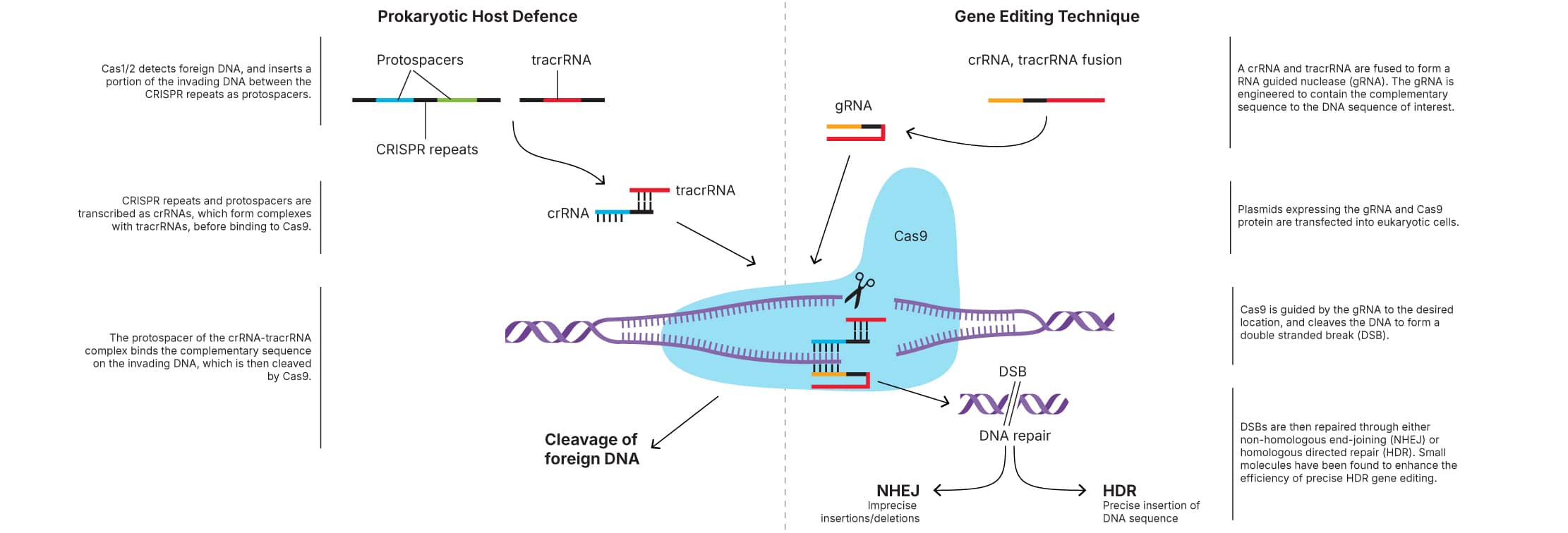
CRISPR (clustered regularly interspaced short palindromic repeats) is a technology for the targeted editing and regulation of genes that can be applied to a number of biological systems.
CRISPR-Cas9 is an RNA-guided DNA endonuclease enzyme (MW: 158.4 kDa), and is part of an acquired bacterial immune response against invading plasmids and viruses. The CRISPR locus is composed of Cas (CRISPR associated) genes, a leader sequence and repeat spacer arrays (CRISPR repeats). The Cas genes encode nucleases that are capable of cleaving DNA. In type II CRISPR systems, invading DNA is detected by Cas1/2, which inserts a portion of the invading DNA into the cell's CRISPR array, between the CRISPR repeats, as a protospacer. The CRISPR array is then transcribed to form crRNA, which forms a complex with transactivating (tra)crRNA and Cas9. The protospacer section of the crRNA then binds to the complementary sequence on the invading DNA, which is cleaved by Cas9.
The CRISPR system has been adapted to create a technique capable of targeted gene editing at specific locations in the genome. By creating double-stranded breaks, CRISPR-Cas9 technology can be used to induce insertion or deletion mutations, specific sequence replacements or insertions, and large deletions or genomic rearrangements at any desired genomic location.
This is made possible by designing RNA guided nucleases. In their simplest form, Cas9 is complexed with a guide RNA (gRNA), which consists of a crRNA and tracrRNA. The gRNA contains a complementary sequence to the DNA sequence of interest, and therefore guides Cas9 to the desired location. Cas9 then cleaves the DNA to create a DSB.
DSBs can be repaired by endogenous non-homologous end-joining (NHEJ) and homology-directed repair (HDR). CRISPR-Cas9-mediated genome editing through NHEJ is imprecise but can reach efficiencies of 20-60%, whereas gene editing efficiency by HDR is more precise but has only been shown to reach 0.5-20%. As HDR is more precise than NHEJ, improving the efficiency of HDR is vital for the use of the CRISPR-Cas9 system in many applications.
Recent research has demonstrated that small molecules can enhance the efficiency of HDR-mediated gene editing. The β3 adrenergic receptor partial agonist L-755,507 has been shown to enhance the efficiency of HDR-mediated GFP insertion and point mutations by 3-fold and 9-fold, respectively in pluripotent stem cells. The small molecule SCR7 pyrazine has also been shown to enhance HDR-efficiency, enhancing insertion of short DNA fragments by up to 19-fold. Small molecules that enhance HDR efficiency could be integral to the application of the CRISPR-cas9 gene editing system.
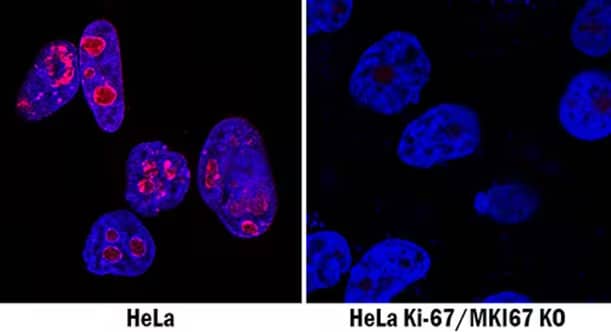
The use of genetic negative controls has been proposed by the International Working Group for Antibody Validation as one of five key criteria for ensuring antibody specificity. Specificity, selectivity and fidelity of antibody-target interactions often varies based on the intricacies of specific analytical applications (e.g., immunoblot vs. immunostaining). Double knockout cell-lysates (Western Blot) and cell-lines (Immunocytochemistry) are the best negative controls to quickly and confidently prove an antibody’s specificity for a target of interest and ensure reproducibility.
Application of CRISPR/Cas9 gene-editing technology for the generation of knockout cell lysates and cell lines ensures the development of useful tools to validate antibody specificity and elucidate gene function. For example in In western blot analysis, an initial indicator of antibody specificity is the presence of a single band at the expected target size, however proper antibody validation requires the use of adequate controls. Because CRISPR/Cas9 gene-editing technology enables complete removal or "knock out" of both alleles of the gene encoding the target protein, antibody specificity is confirmed by demonstrating that a protein band is only present in the wildtype and not the KO cell lysate in WB analysis.
NOTE: To confirm differential expression, it is also important to analyze a separate target protein as a loading control (e.g., GAPDH) to demonstrate that all samples were loaded and transferred equally across wells.