Analysis of Monoclonal Antibodies with Carboxypeptidase B by Maurice
Introduction
The heavy chain of human IgG contains a C-terminal lysine residue which is typically cleaved off by carboxypeptidase in vivo1 . For decades, it was believed that the C-terminal clipping of lysine residues did not affect antibody structure and function, and was therefore not considered a critical quality attribute (CQA)2 . However, the significance of C-terminal lysine clipping is just now being recognized, with a recent study demonstrating that antibodies that retain the C-terminal lysine are attenuated for complement-dependent cytotoxicity3. Thus, assessing C-terminal lysine variants is required in monoclonal antibody (mAb) process development.
Cleaving C-terminal lysine residues in vitro with carboxypeptidase B (CpB) is relatively straightforward, but there are some factors that can confound results. For example, the components in the formulation buffer can inhibit CpB activity. To provide consistent results, these inhibitors should be removed with a desalting column. Furthermore, many pI markers are peptides, and several basic peptide pI markers have C-terminal lysine or arginine residues. Thus, residual CpB can digest these markers, resulting in the loss of pI standards and the possible addition of unwanted lower pI peaks. This makes it difficult to calibrate the apparent pI of your target molecule. In such events, cooling the ampholyte and sample to 4 °C prior to mixing can be useful for the retention of pI calibration markers. Also, citric acid is a known inhibitor of CpB activity4, and its addition can help retain the pI markers.
To help you navigate potential pitfalls in CpB analysis of antibodies, we provide a protocol to optimize CpB treatment prior to analysis on Maurice™. Using adalimumab as an example, we demonstrate that desalting the formulation buffer is required for optimal CpB activity, resulting in the successful removal of the C-terminal lysine peaks. Furthermore, preservation of the pI markers can be achieved by cooling the ampholyte solution and digested sample before mixing, and by the addition of citric acid to inhibit residual CpB activity.
Materials
ProteinSimple
- Instrument: Maurice
- Maurice cIEF Method Development Kit (PN PS-MDK01-C)
- Maurice cIEF Cartridges (PN PS-MC02-C)
Other reagents
- Carboxypeptidase B (Sigma-Aldrich, PN C9584)
- Amicon Ultra 0.5 mL centrifugal filters (Sigma-Aldrich, PN Z677108)
- Citric acid (Sigma-Aldrich, PN C1909) prepared as a 400 mM solution
Sample
- •Adalimumab (1 mg/mL)
Methods
- Reconstitute CpB in water to a concentration of 1 mg/mL. Aliquot CpB and freeze at -80 °C for future use. Thaw aliquots on ice before use.
- Add CpB at a ratio of 1:100 (CpB to sample). For example, add 1 μL CpB to 100 μL of sample.
- Vortex gently and incubate at 37 °C for 30 minutes.
- Add the CpB-treated sample to ampholyte mix and briefly vortex. For example, add 40 μL of treated sample to 160 μL of ampholyte mix.
- Analyze the sample on Maurice or iCE3.
NOTE: If the lysine-containing peaks remain, desalt the sample prior to CpB digestion with Amicon Ultra 0.5 mL centrifugal filters according to the manufacturer's instructions.
NOTE: If you observe loss of the pI markers, take the following steps immediately after CpB digestion:
- Add citric acid at a ratio of 1:100 citric acid to sample to achieve a 4 mM final concentration and gently vortex. Incubate at room temperature for 20 minutes.
- Individually cool the sample and the ampholyte solution to 4 °C prior to mixing the two. The final citric acid concentration in the sample and ampholyte mix will be 0.8 mM.
Results
Following the protocol described above, we treated adalimumab with CpB prior to icIEF analysis. Compared to the no-treatment control, two peaks corresponding to lysine variants were removed by CpB, revealing new basic variants of adalimumab that were otherwise undetected (Figure 1).
There were two challenges that had to be overcome to arrive at the results in Figure 1. First, inhibitors in the formulation buffer prevented CpB activity, so desalting the sample prior to CpB treatment was required. To demonstrate this, we set up three samples: (1) CpB added to the sample after desalting the formulation buffer, (2) CpB added directly in the formulation buffer without desalting, and (3) a negative control sample with no CpB added. From these results, only desalting adalimumab before digestion successfully removed the C-terminal lysine peaks (Figure 2).
The second challenge that we encountered was collateral digestion of the peptide markers. The desalted sample and its ampholytes in Figure 2 were cooled to 4 °C prior to mixing, whereas the other two samples (undesalted and untreated) and their respective ampholyte mixes were kept at room temperature. To show the effect of precooling the sample and the ampholyte mix on the basic pI marker, Figure 3 shows the same electropherograms overlaid with each other. The inset is a magnification of the basic pI marker. This analysis reveals that the sample cooled to 4 °C retained more of the basic pI marker than the sample that was kept at room temperature, indicating that cooling the sample can reduce unwanted residual CpB activity that can digest the peptide standards. Citric acid may also be added to the sample at a 4 mM final concentration immediately following CpB digestion to reduce undesirable CpB activity as described above. It should be noted that adding citric acid may impact the pH gradient by increasing the anodic zone and pushing peaks toward the cathodic end.
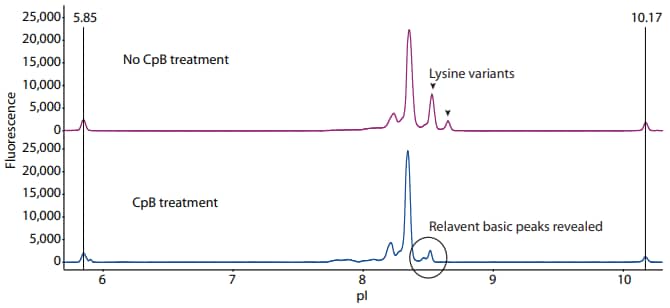
Figure 1. Treatment of adalimumab with CpB removes the lysine variants and reveals new relevant basic variants by icIEF analysis. The fluorescence exposure time is 8 seconds
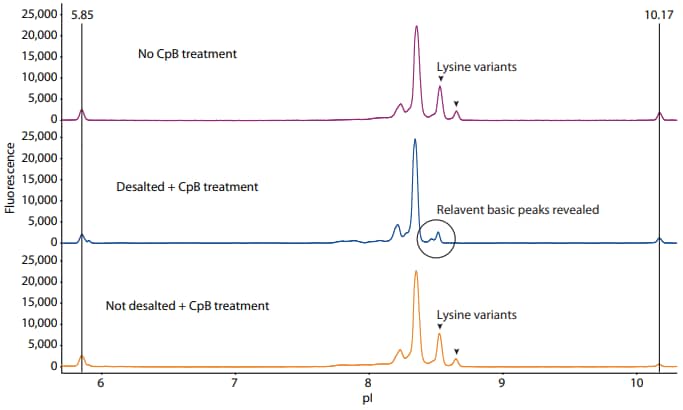
Figure 2. Activity of CpB when added after desalting the formulation buffer (middle panel), added directly to the formulation buffer without desalting (bottom panel), or not added at all (top panel). The fluorescence exposure time is 8 seconds.
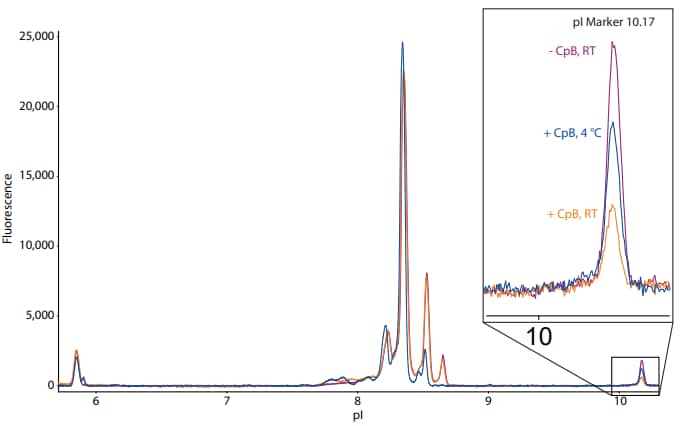
Figure 3. Collateral digestion of basic pI marker is reduced when the sample and ampholyte mix are cooled to 4 °C. The inset is a magnification of the basic pI marker of the desalted sample, which was cooled to 4 °C following CpB digestion, and the undesalted and untreated samples, which were kept at room temperature (RT). The fluorescence exposure time is 8 seconds.
Conclusion
CpB analysis of mAbs commonly encounters two challenges: (1) inhibitors in the formulation buffer that prevent CpB activity, and (2) unwanted collateral digestion of terminal lysine and arginine residues in the peptide standards. Here, we describe steps that can be taken to effectively overcome these challenges, resulting in robust charge separation and analysis of the lysine variants of a monoclonal antibody on Maurice or iCE3.
References
- C-terminal lysine processing of human immunoglobulin G2 heavy chain in vivo, B Cai, H Pan and G Flynn, Biotech Bioeng, 2011; 108(2):404–12.
- Microheterogeneity of recombinant antibodies: analytics and functional impact, B Beyer, M Schuster, A Jungbauer and N Lingg, Biotechnol J, 2018; 13:1–11.
- Human IgG is produced in a pro-form that requires clipping of C-terminal lysines for maximal complement activation, E van den Bremer, F Beurskens, M Voorhorst, P Engelberts, R de Jong, B van der Boom, E Cook, M Lindorfer, R Taylor, P van Berkel and P Parren, mAbs, 2015; 7:672–80.
- The kinetics of carboxypeptidase B activity, E Wolff, E Schirmer and J Folk, J Biol Chem, 1962; 237:3094–9.