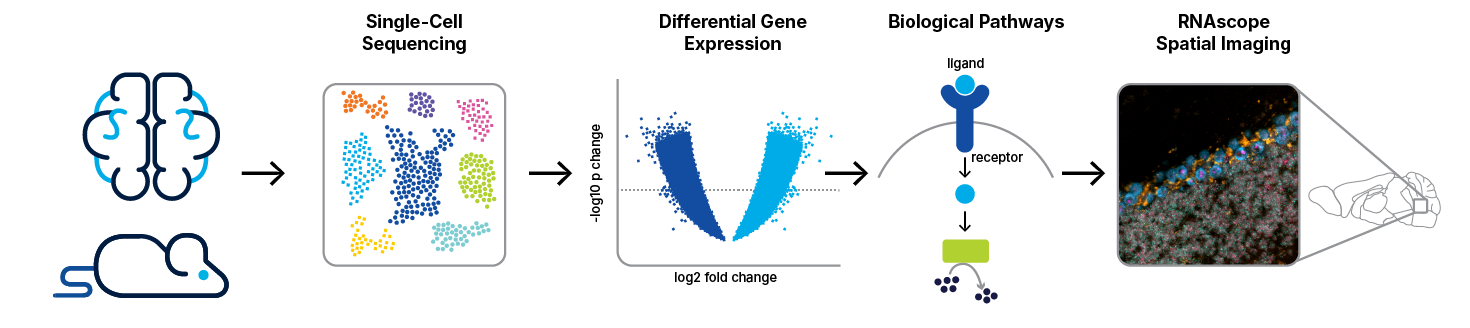
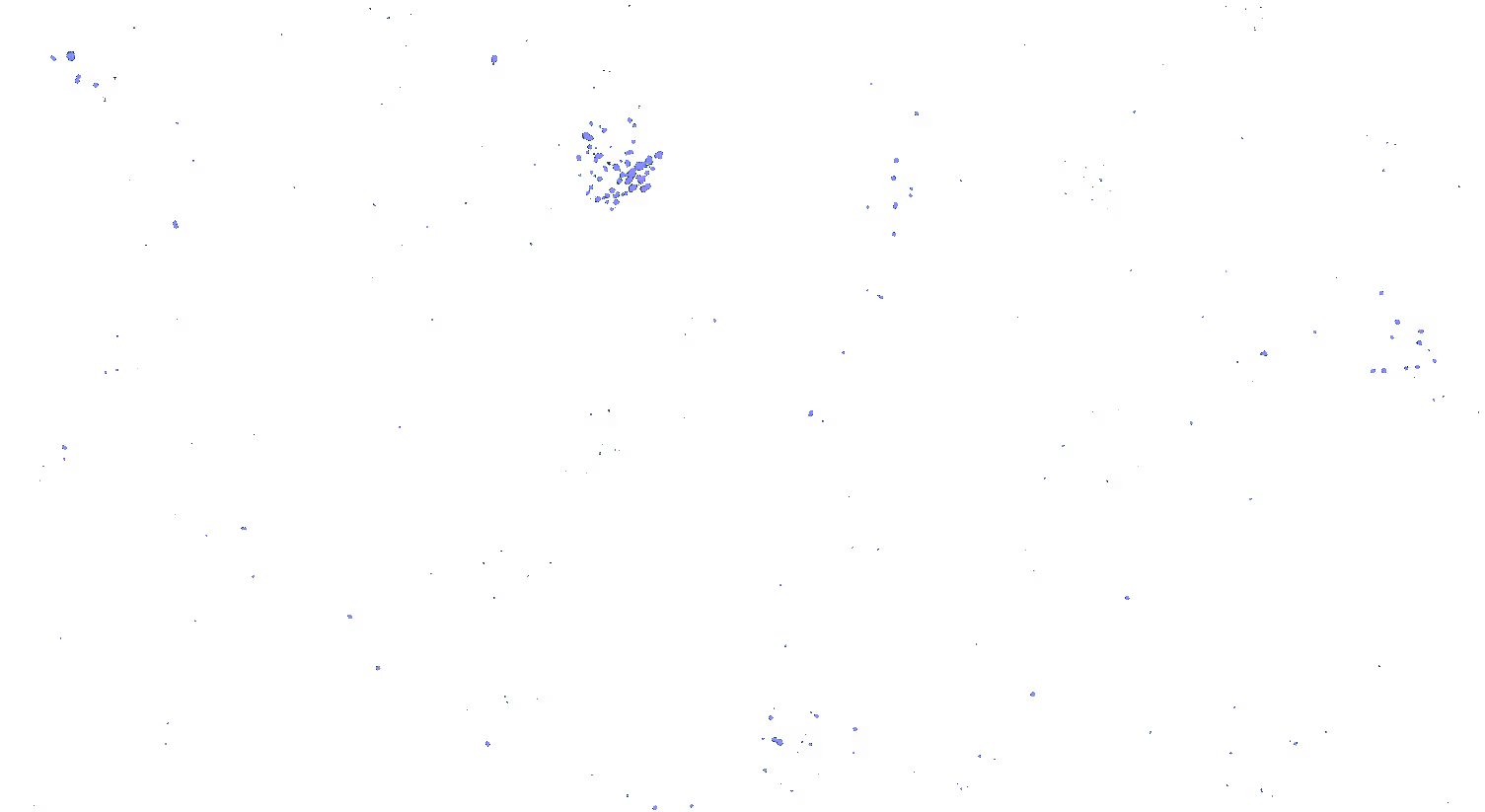
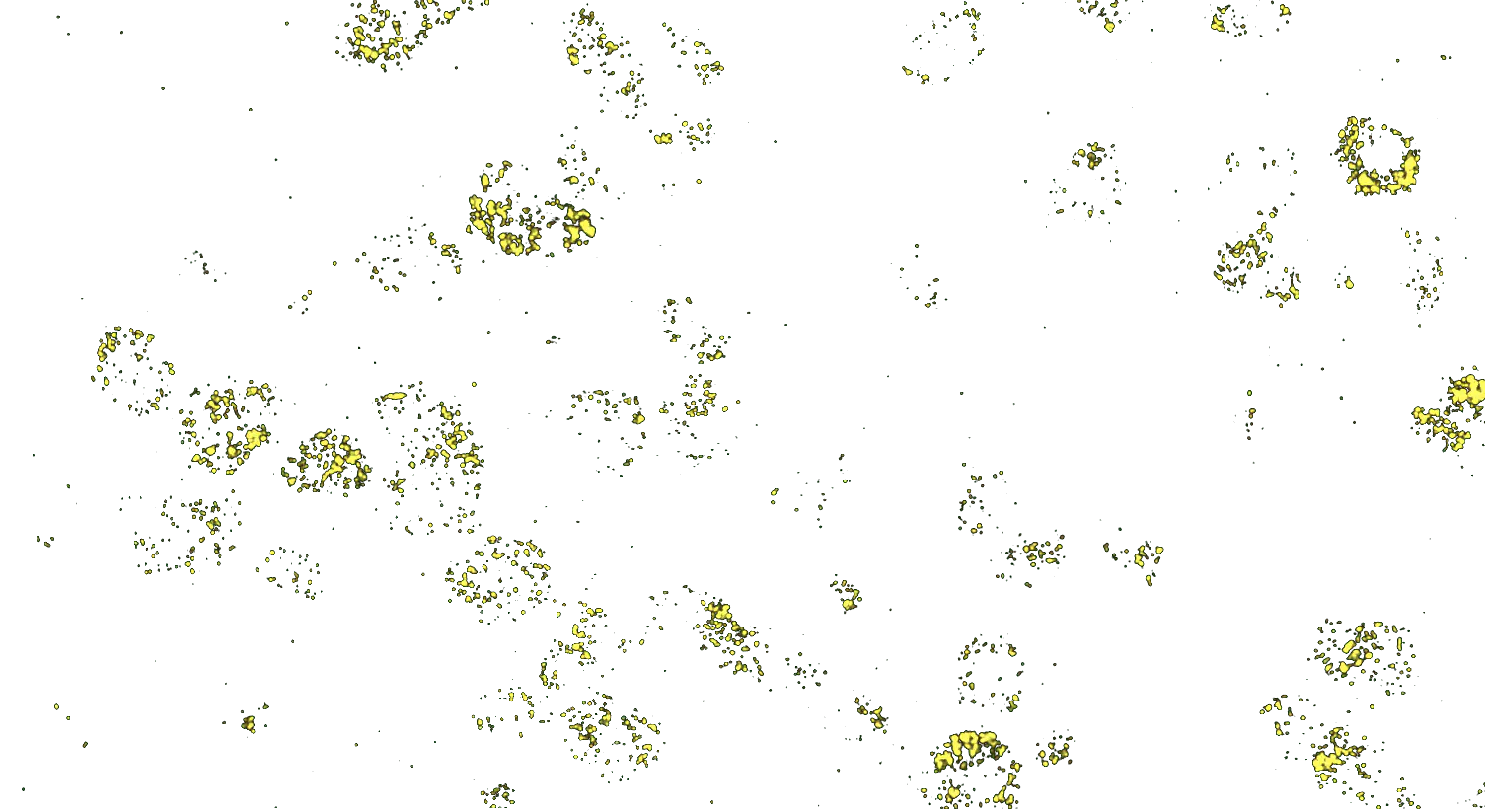
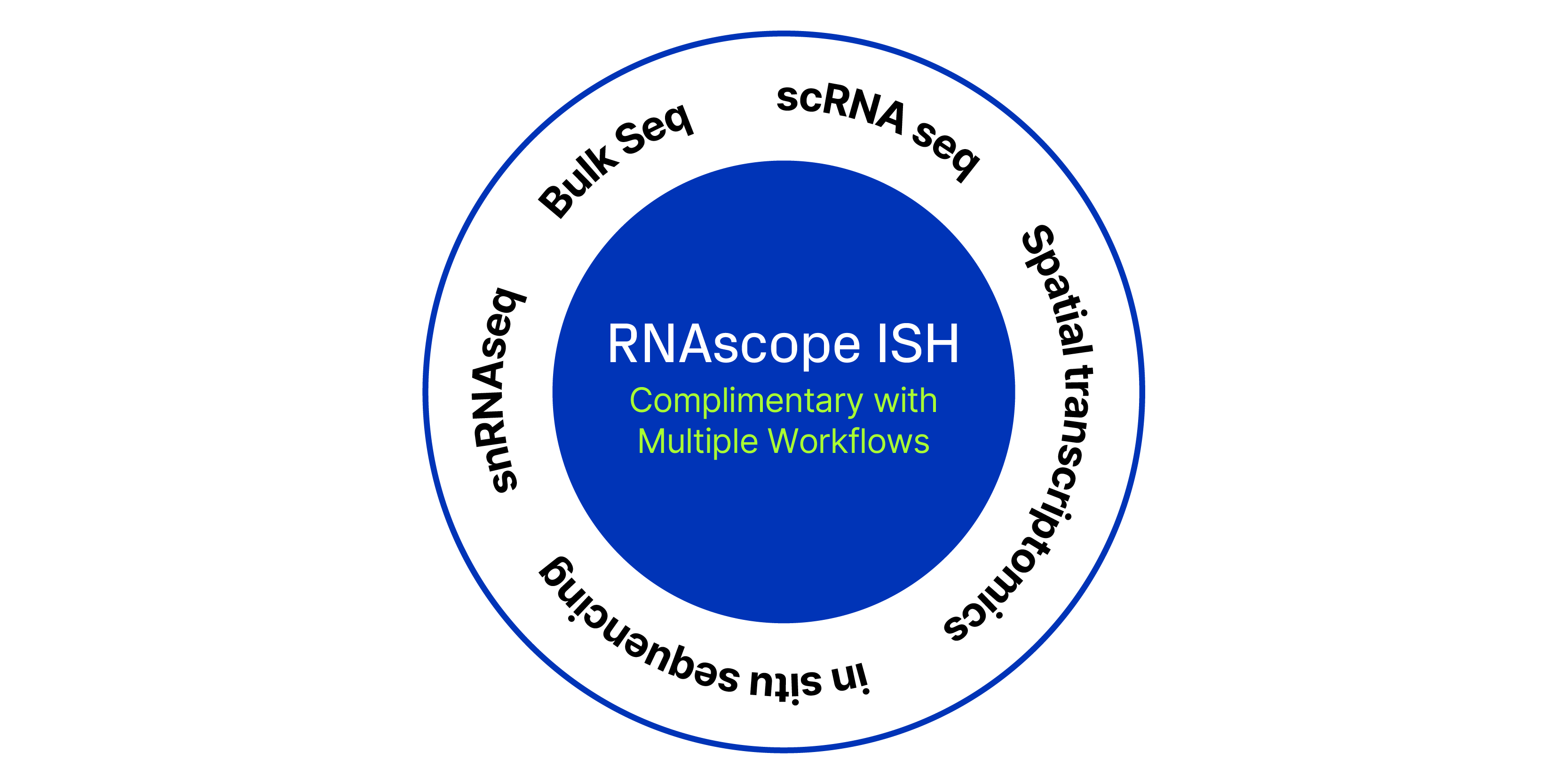
Transcriptomic technologies such as single cell RNA sequencing (scRNA-Seq), single-nuclei, or bulk-sequencing have revolutionized the study of cellular diversity, enabling researchers to identify known and novel cell types and gain insights into tissue structure and function.
However, such transcriptomic methodologies require cell dissociation, which disrupts the spatial context of the cells. To address this limitation, it is crucial to pair molecular information with spatially-resolved methods like RNA in situ hybridization (ISH). Using RNAscope™ ISH allows for the visualization of gene expression within the intact tissue, preserving the spatial organization and providing a more comprehensive understanding of cellular functions, individual cell profiles, and their spatial arrangements, enhancing our ability to study and interpret complex biological systems.
Spatial Mapping
Spatial Mapping
Validate the presence, abundance, and spatial localization of novel cell subtypes and cell markers.
Cellular Dynamics
Cellular Dynamics
Identify meaningful changes in cell phenotypes and cell-cell interactions in human pathology and animal models.
Activation States
Activation States
Visualize changes in single-cell function including initiation of signaling pathways and transcriptional activation.
Validate Gene Signatures with Spatial Context at the Single Cell Level
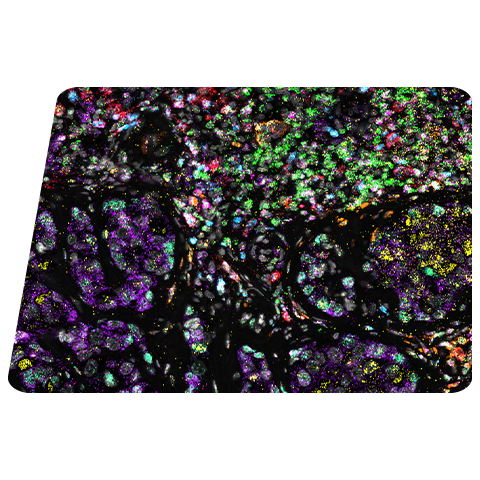
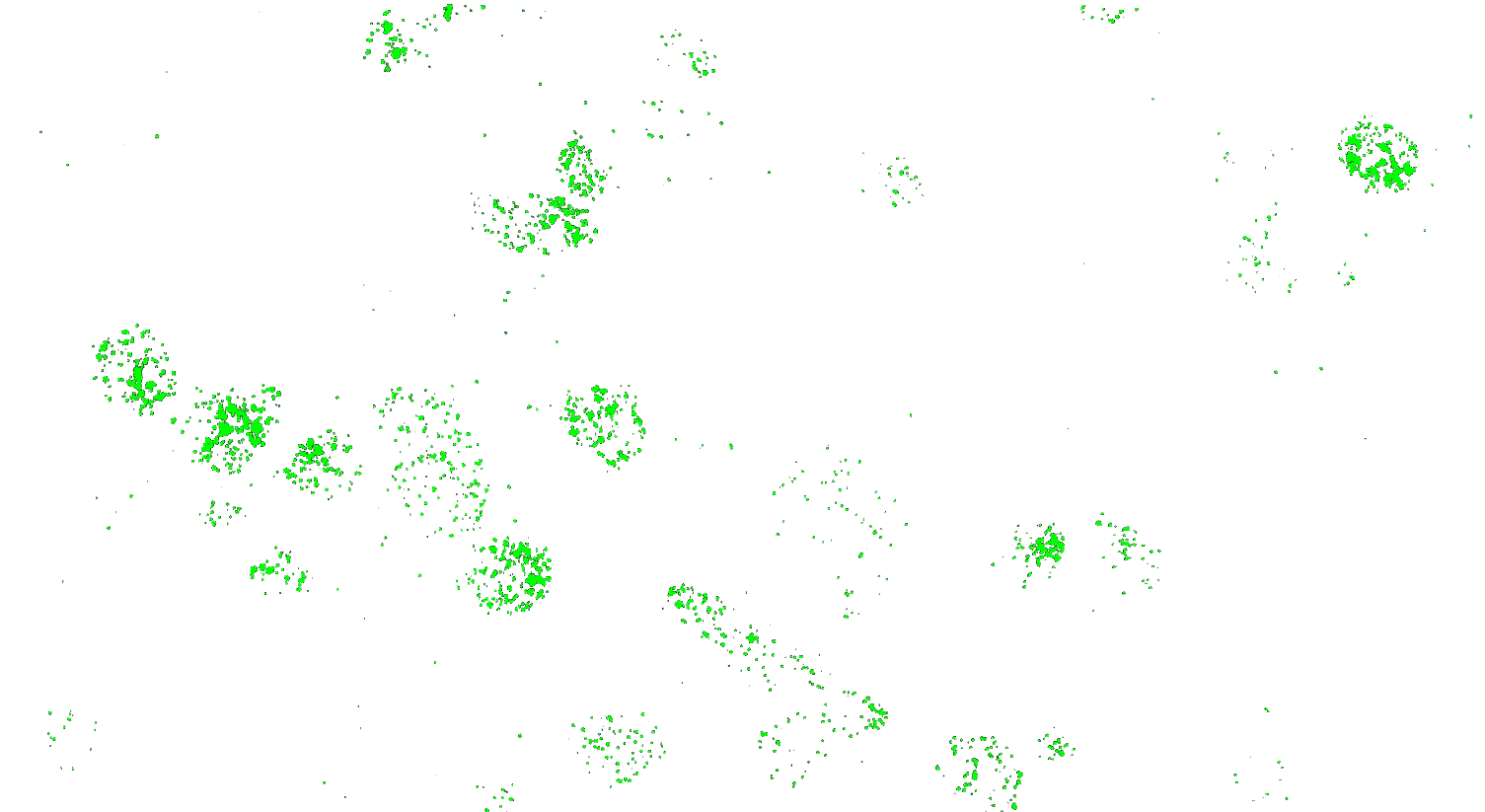
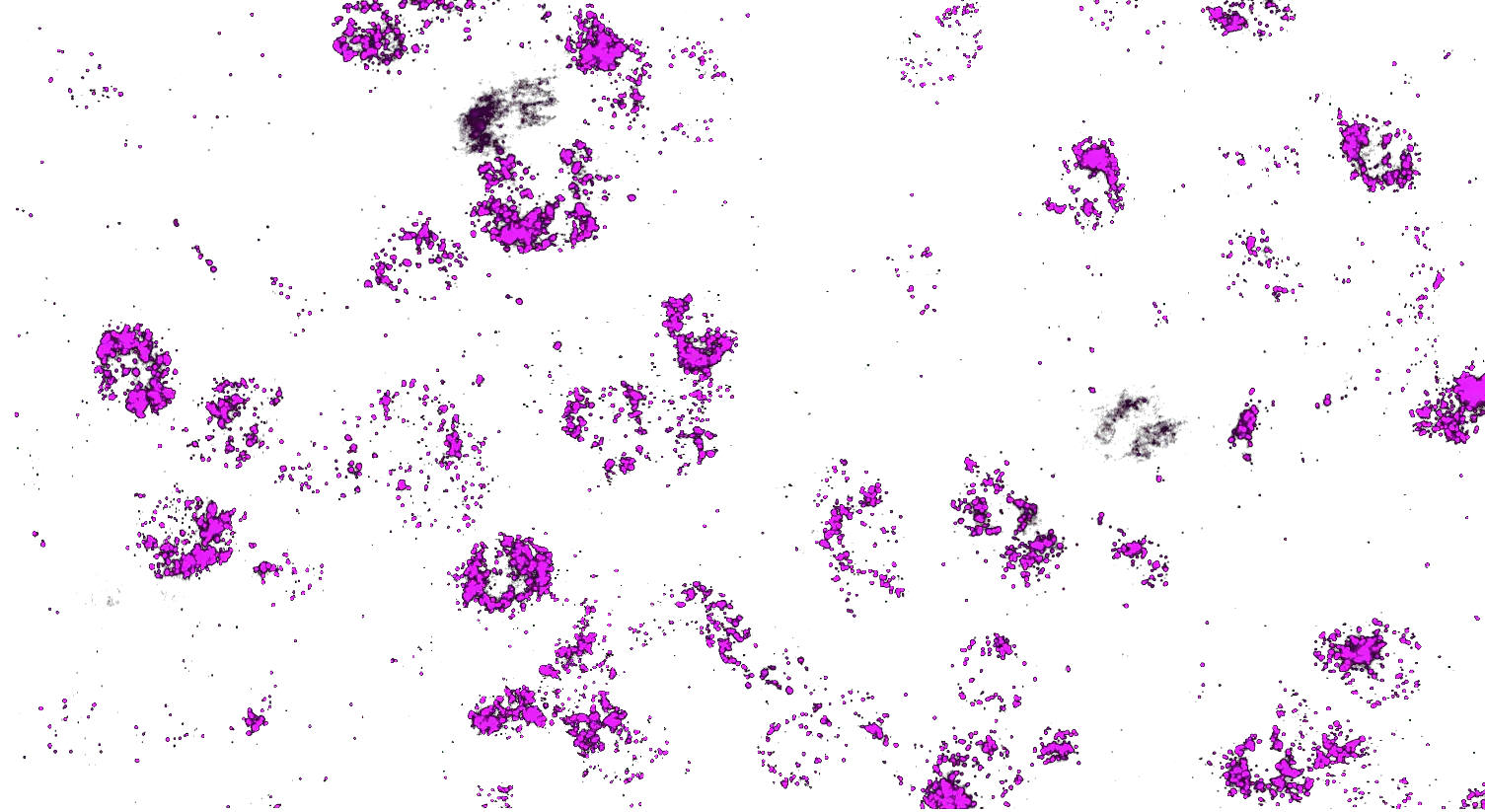
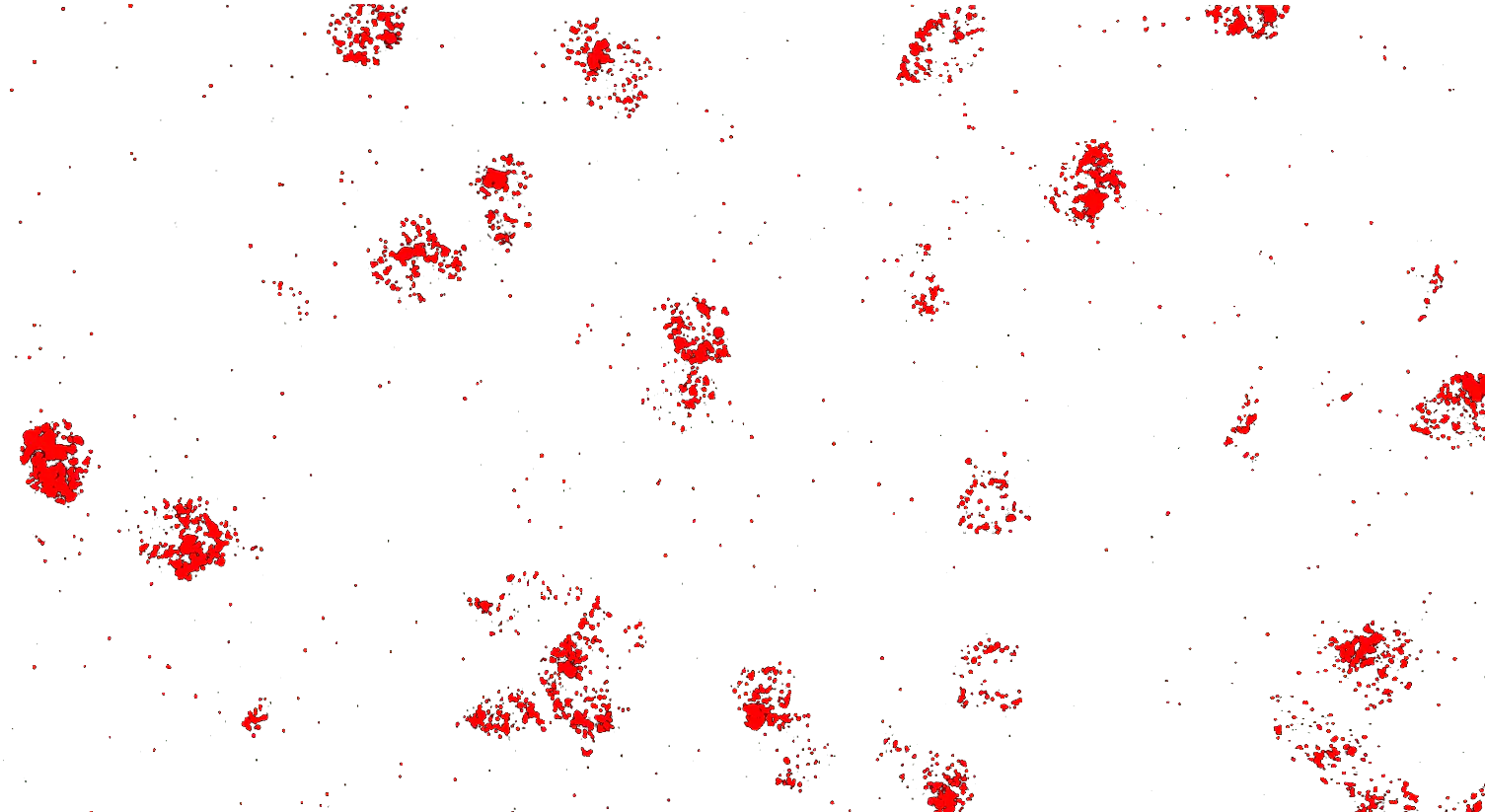
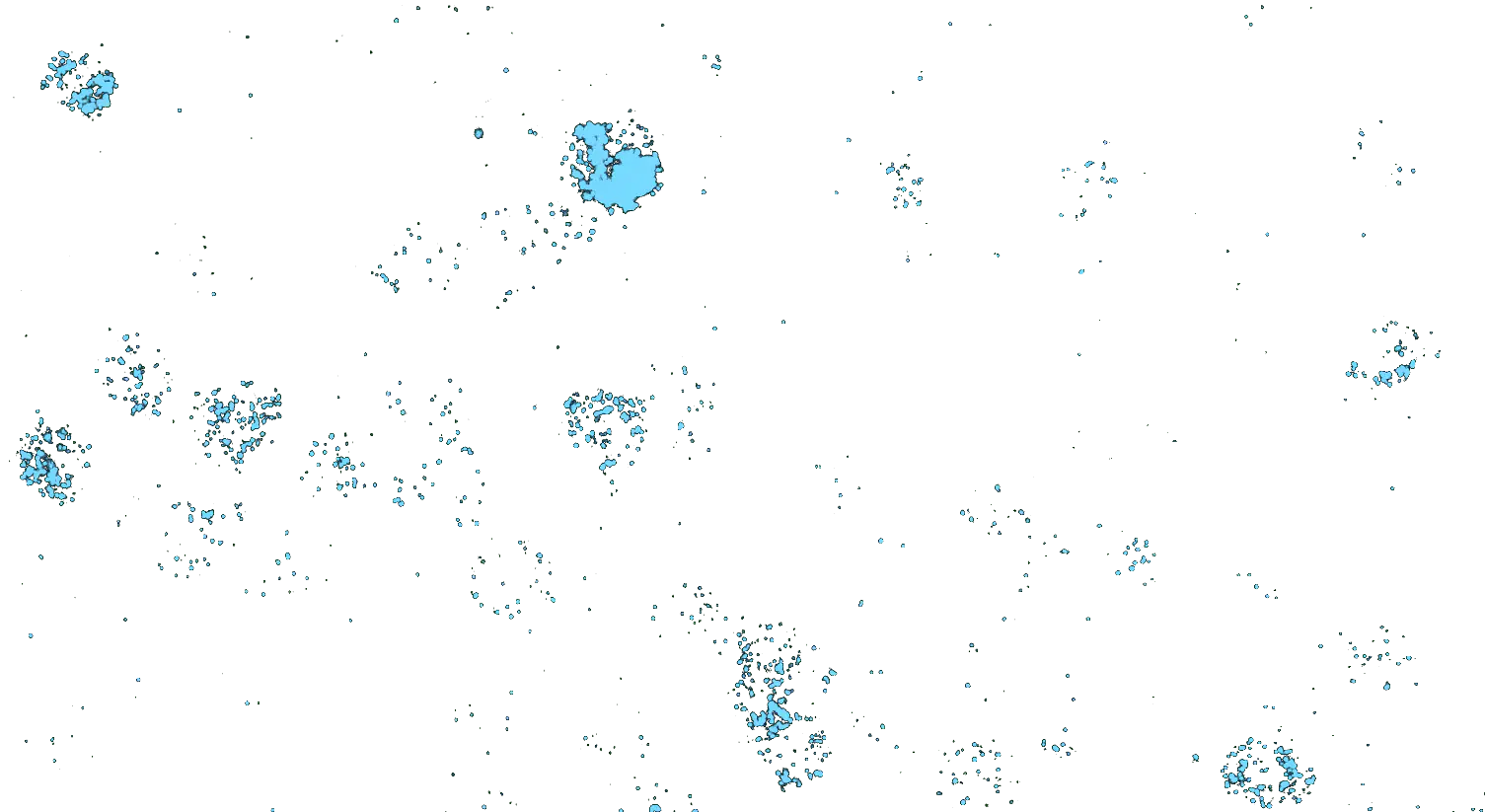
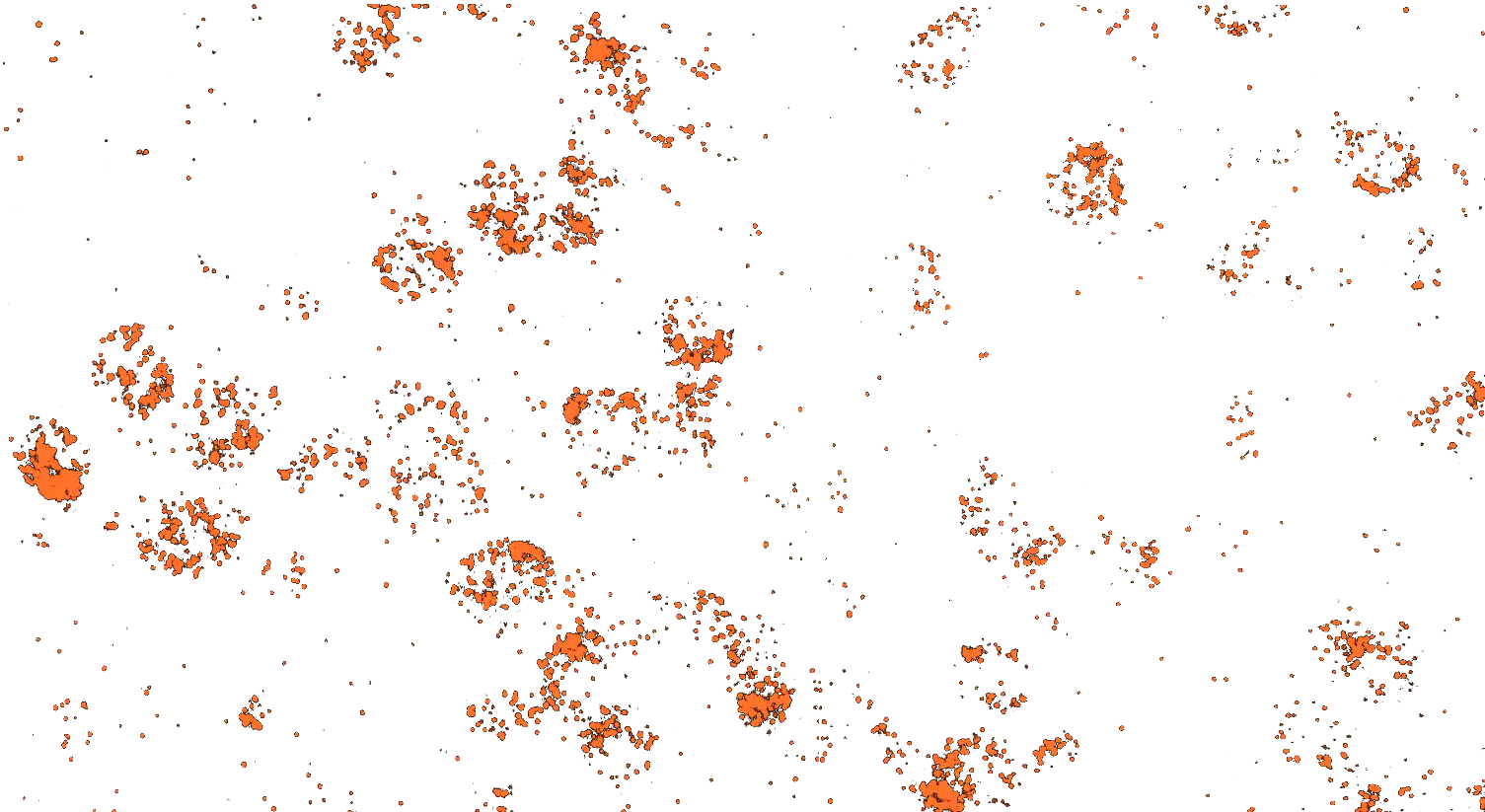
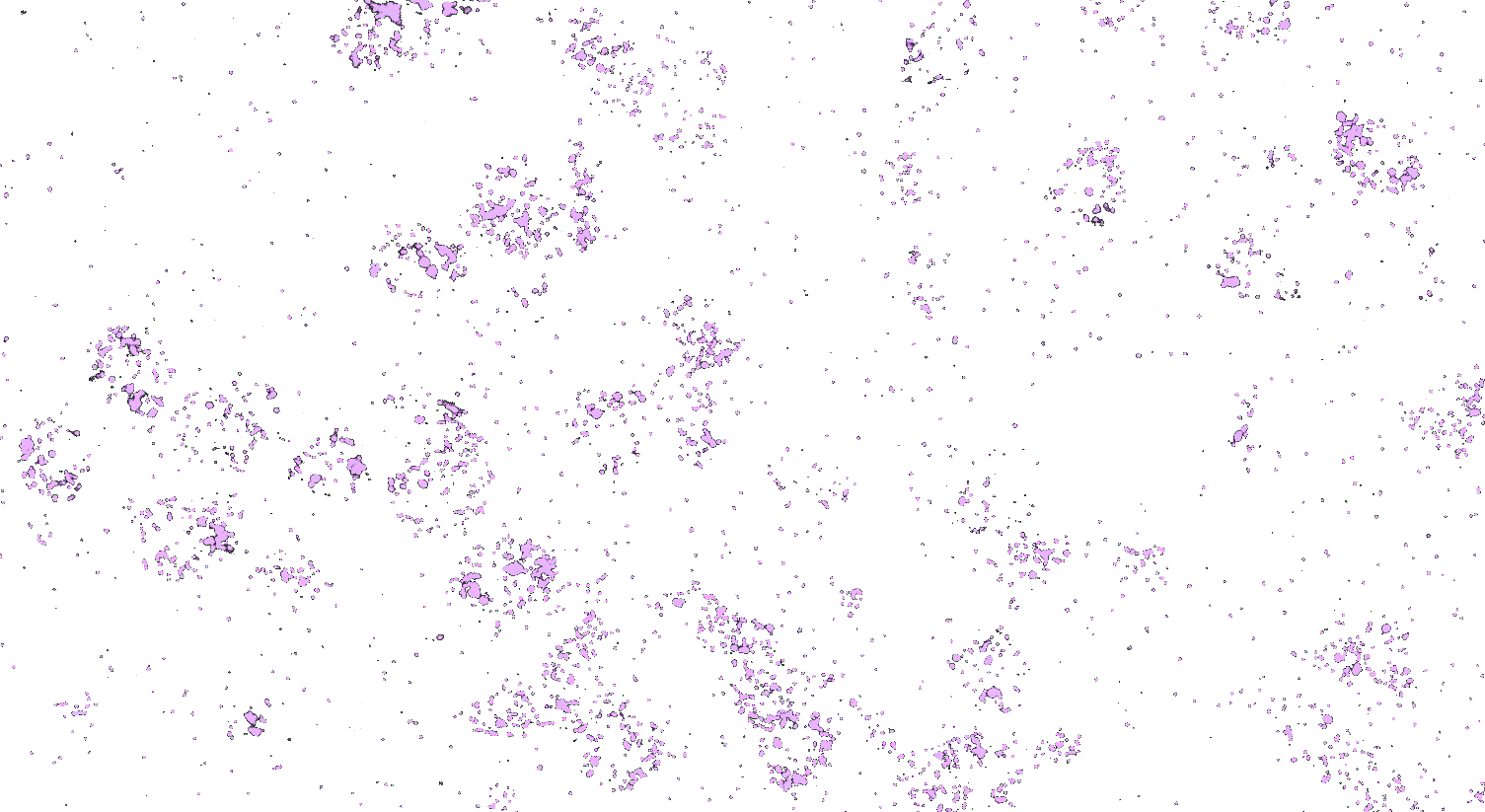
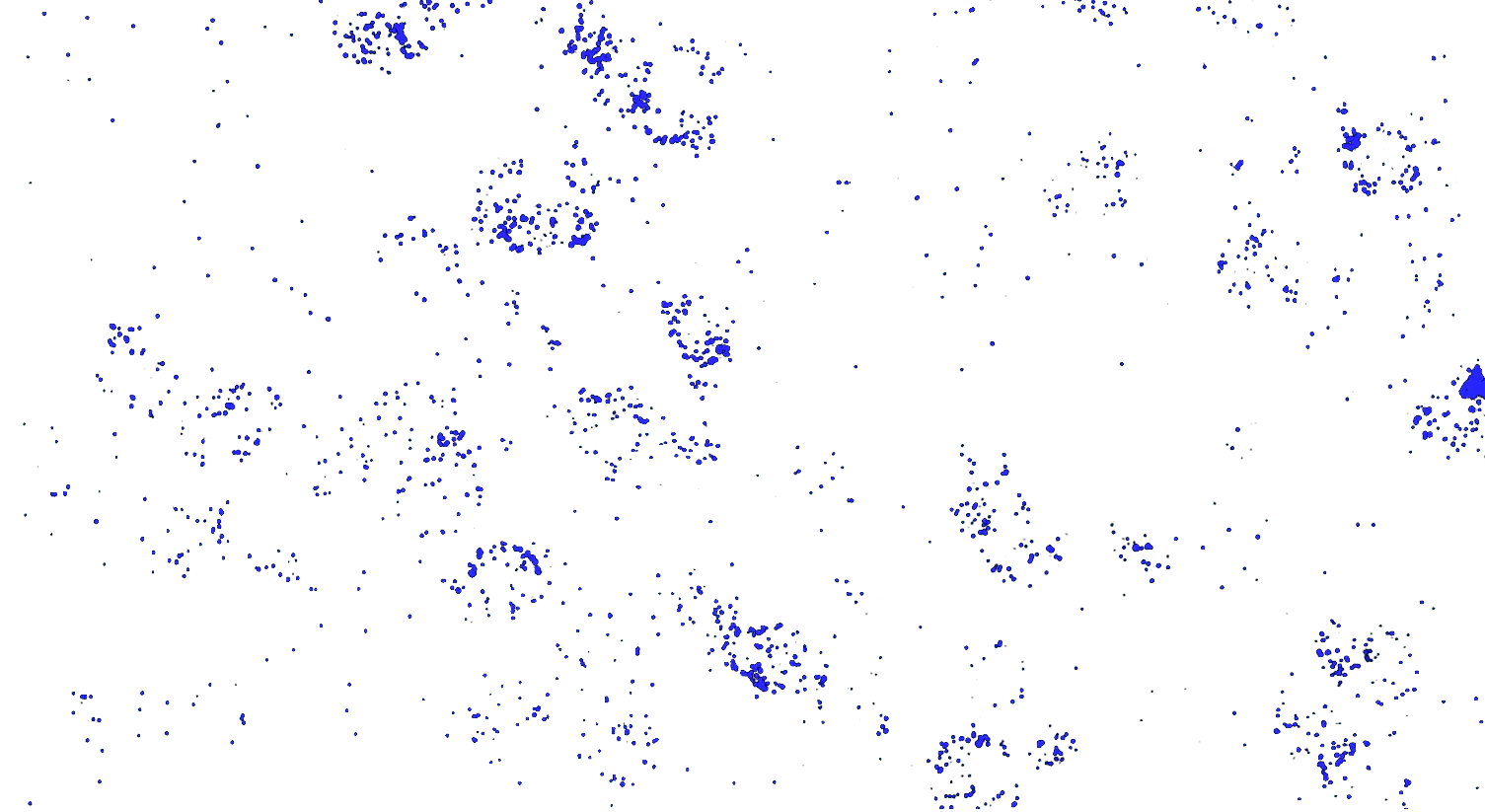
Visualizing striatal Drd1a and Drd2 medium spiny neurons (MSNs) using the RNAscope HiPlex assay for 12 targets in fresh frozen mouse brain sections.
Targets are Drd1a, Drd2, Foxp1, Pcdh8, Synpr, Htr7, Meis, Calb1, Crym, Cnr1, Wfs1, and TH.
Getting Started is Easy
1. Easy Probe Selection
Select just the genes you want from an extensive online catalog and custom design services.
2. No Spatial Instrument Required
Get results quickly using RNAscope manual kits and your own fluorescent microscope. Easily scale with automated protocols.
3. Expert Technical Support
Our experienced technical specialists in RNA spatial imaging will help you get up and running quickly.
Customer Success Stories
Webinar: Spatial Organization of Multicellular Immune Hubs in Human Colorectal Cancer
In this webinar, Dr Jon Chen, Massachusetts General Hospital Cancer Center, Harvard Medical School, will discuss this study in greater detail and demonstrate a path to discovering multicellular interaction networks that underlie immunologic and tumorigenic processes in human cancer.
Learning Objectives:
- How scRNA-seq can be used to reveal shared and distinct features of human MMRd and MMRp colorectal cancer
- How multicellular immune hubs can be predicted based on co-variation of gene programs
- Why understanding the molecular mechanisms underlying immune hubs and their temporal and spatial regulation upon treatment is critical for advancing cancer therapy