Viral Pathogens
Classification of Viruses
Viruses are "obligatory intracellular parasites" that subvert the molecular machinery of host cells to replicate. Classification of viruses is based on several criteria:
Morphology- Viruses consist of a core of nucleic acid material which constitutes their genome and a protein-based coating or capsid. An additionally lipoprotein bilayer surrounds some viruses and is the basis for their classification as enveloped and non-enveloped viruses. Based on capsid shape, viruses may be classified as filamentous or helical (e.g., plant viruses-TMV and animal viruses such as rabies virus), icosahedral (e.g., human pathogens such as adenoviruses and Hepatitis A virus), and complex such as “head and tail viruses” (e.g., bacteriophages) and pleomorphic viruses (e.g., Ebola virus).
Genomic composition- The viral genome together with surrounding proteins constitutes the nucleocapsid. Viral genetic material may be encoded by RNA or DNA which may be single (ss) or double stranded (ds). The genomic material in ssRNA viruses may consist of a positive sense or negative sense strand.
Replication Mechanism- Depending on the nature of their genome, viruses rely on different mechanisms for mRNA synthesis. The Baltimore classification, conceived by and named after virologist David Baltimore, classifies viruses based on their genome composition and replication mechanisms. Seven different groups are recognized under this system: Group 1- Double stranded DNA, Group 2- Single stranded (+) sense DNA, Group 3- Double stranded RNA, Group 4- Single stranded (+) sense RNA, Group 5- Single stranded (-) sense stranded RNA-, Group 6- Single stranded (+) sense RNA with DNA intermediate- (Retro-transcribing), Group 7- Double stranded DNA with RNA intermediate (Retro-transcribing).
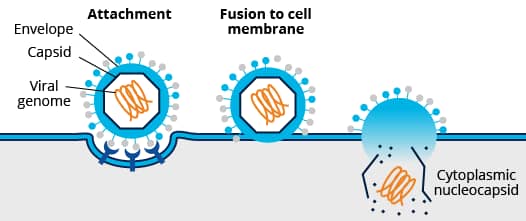
Viral Entry
Mechanisms of cell entry differ between enveloped and non-enveloped viruses. For cell entry, the lipid bilayer of enveloped viruses fuses directly with the host cell’s plasma membrane or endosomal membrane. This process is mediated by the interaction of envelope viral proteins with key cellular receptors. Fusion via the endocytic compartment is a common mechanism for host cell entry used by a variety of viruses including alphaviruses (e.g., mosquito-borne chikungunya virus), flaviviruses (e.g., dengue and zika viruses), orthomyxoviruses (e.g., influenza viruses), and rhabdoviruses (e.g., rabies virus). In contrast, non-enveloped viruses rely on capsid proteins which bridge the plasma or endosomal membrane (e.g., pore formation) allowing viral genome entry. In addition to endosomal entry, non-enveloped viruses may bridge other internal cellular membranes including the Golgi (e.g., papillomavirus) and ER (e.g., SV40).
Non-enveloped vs Enveloped Viruses
Immunocytochemical (ICC) staining of cells infected with adenovirus using Adenovirus Mouse Monoclonal Antibody (M58 + M73) [Catalog # NB120-3648].
Immunocytochemical (ICC) staining of cells infected with HHV8 using an ORF73/HHV8 Rat Monoclonal Antibody (LN53). [Catalog # NBP1-22767]
Viral Attachment Factors and Entry Receptors
Initial viral-cell contact or attachment is mediated by electrostatic interactions between viral proteins and host surface carbohydrate groups (e.g., glycosaminoglycan groups such as heparan sulphate and sialic acid). Binding to specific receptors follows initial weaker interactions with the host cell membrane and leads to viral protein processing and cellular signaling. The receptors engaged by a specific virus determine its internalization path.

Viruses engage different cellular mechanisms to gain entry into host cells, including direct entry at the cell membrane and through endocytic pathways such as clathrin-mediated, caveolae/raft-dependent, clathrin/caveolin-independent, and macropinocytosis. However, examples abound of viruses which exploit more than one mechanism for cell entry, and entry mechanisms may differ based on the host cell type. For SARS coronavirus, a clathrin-dependent entry pathway has been identified, nevertheless SARS coronavirus cell entry may also occur through a clathrin/caveolin-independent pathway.
NOTE: HIV entry mechanisms are cell type-dependent and include clathrin-mediated, dynamin-dependent endocytosis, and macropinocytosis.
Entry Receptors for Major Human Viral Pathogens
Viral host cell entry is a multistep process involving a number of cellular attachment factors, receptors, and proteases. For some viruses, a predominant entry receptor has been identified (e.g., SARS coronavirus ACE-2 receptor). However, identification of a unique entry receptor remains challenging for important human viral pathogens including Hepatitis A Virus (HAV) and Chikungunya virus (CHIKV).
| Virus Family | Virus Species | Enveloped (E) or Non-Enveloped (N) | Genome | Entry Receptors | Main Endocytic Pathways | |
|---|---|---|---|---|---|---|
| Adenovirus | Adenovirus 2 | N | dsDNA | CXADR, alpha 5 integrin | Clathrin-dependent | |
| Adenovirus | Adenovirus 3 | N | dsDNA | CD46, alpha 5 integrin | Macropinocytosis | |
| Coronavirus | MERS-CoV | E | ssRNA+ | DiPeptidyl Peptidae-4 (DPP4) | Clathrin-dependent | |
| Coronavirus | SARS-CoV | E | ssRNA+ | ACE-2 | Clathrin-dependent, Clathrin /caveolin-independent | |
| Filovirus | EBOV | E | ssRNA+ | TIM-1, Axl, NPC-1 | Macropinocytosis | |
| Flavivirus | Dengue | E | ssRNA+ | DC-SIGN, Mannose receptor (MMR), CD14, HSP-70, HSP-90, GRP78 | Clathrin-dependent | |
| Flavivirus | HCV | E | ssRNA+ | CD81, SR-BI, LDLR, Claudin-1, Occludin | Clathrin-dependent | |
| Flavivirus | Zika | E | ssRNA+ | DC-SIGN, TIM-1, TAM (TYRO3, AXL, MER) | Clathrin-dependent, Mucolipin-2-dependent | |
| Orthomyxovirus | Influenza | E | ssRNA+ | Sialic acid |
|
|
| Papovavirus | HPV | E | dsDNA | syndecan-1, laminin-5, integrin alpha 6 | Clathrin-dependent | |
| Picornavirus | HAV | N eHAV-Quasi-enveloped** |
ssRNA+ | TIM-1, ALIX, Integrin beta 1** | Clathrin-dependent, Dynamin-dependent | |
| Picornavirus | Rhinovirus | N | ssRNA+ | ICAM-1, LDLR, p-cadherin | Clathrin-dependent, Clathrin independent, Macropinocytosis | |
| Retrovirus | HIV | E | ssRNA+ | Clathrin-dependent, Dynamin-dependent, Macropinocytosis | ||
| *Togavirus | Chikungunya | E | ssRNA+ | TIM-1, DC-SIGN | Clathrin-dependent |
*Yeast two-hybrid screens and loss of function screens have identified other interacting proteins including Actin gamma 1, collagen type I-alpha-2, Tyrosine phosphatase non-receptor 2 (PTPN2), Fuzzy homolog, Tspan9, PHB-1 (prohibitin 1), Mxra-8. The role of these proteins in viral entry remains inconclusive.
**Quasi-enveloped HAV virions are secreted from infected cells through a non-lytical pathway and are surrounded by host cell derived membranes. A specific HAV cell entry receptor remains elusive, proteins listed participate in viral attachment and endosomal trafficking.
Identifying Viral Entry Mechanisms
Elucidating viral entry mechanisms often relies on the use of small molecule inhibitors targeting specific molecules or events in the endocytic pathway. Small molecule inhibitors present various advantages including ease of acute application and potential reversibility of effects. A main disadvantage to chemical manipulation of endocytic pathways is the potential for generalized disruption of endocytosis or other cellular processes. Therefore, combined use of chemical inhibitors with other approaches such as targeted genetic manipulation of endocytic relevant targets is recommended.
| Endocytic Pathway | Inhibitors | Activities | Notes |
|---|---|---|---|
| Caveolin-dependent endocytosis | Filipin III | Binds sterols with high affinity, complexes with membrane cholesterol | Affects cholesterol dependent processes and is cytotoxic, inhibits clathrin-mediated endocytosis |
| Okadaic acid | Inhibitor of phosphatase 1 and 2A, promotes removal of caveolae from plasma membrane | Inhibits clathrin-mediated endocytosis | |
| Phorbol 12-myristate 13-acetate | PKC activator, inhibits caveolae dependent entry of Ebola virus | Low toxicity, interferes with endocytic traffic | |
| Macropinocytosis | Amiloride | Inhibitor of Na+/H+ exchange at cell membrane | Blocks clathrin-mediated and lipid raft-dependent endocytosis |
| Cytochalasin D | Inhibitor of actin polymerization | Blocks phagocytosis, affects several endocytic pathways | |
| Latrunculin A, Latrunculin B | Inhibitor of actin polymerization | Blocks phagocytosis, affects several endocytic pathways | |
| Clathrin-mediated endocytosis | Dynasore | Inhibitor of dynamin GTPase activity | Affects cholesterol homeostasis, disrupts lipid rafts, and destabilizes actin |
| Cytochalasin B | Inhibitor of actin polymerization | Affects several endocytic pathways |
What are Neutralizing Antibodies?
During an adaptive immune response against viral pathogens or following vaccination, generation of neutralizing antibodies represents the best indicator of protection. Neutralizing antibodies by definition are those able to target specific surface viral antigens and block the virus replication cycle at an early stage, prior to transcription and translation events. Viral neutralization may occur through different mechanisms including inhibition of attachment, receptor or co-receptor binding, and blockade of viral-cellular membrane fusion or cell membrane penetration. Neutralization events may occur by inhibition of interactions at the host cell surface or within the endosomal compartment. Additionally, some neutralizing antibodies may induce viral aggregation leading to reduced infectivity.
| Neutralizing Antibody | Role of Targeted Protein |
|---|---|
| Chikungunya (CHIKV) (DDX9100P-100, DDX9102P-100, DDX9103P-100, DDX9104P-100) | Envelope glycoproteins E1 (interacts with cellular receptor to mediate viral entry via membrane fusion) and E2 (involved in attachment to receptor) are encoded by a large polyprotein (capsid, E3, E2, 6K,E1). |
| Flavivirus group antigen (NBP2-52666) | E protein (Dengue virus, West Nile Virus, Japanese Encephalitis, Yellow Fever Virus, Zika virus) is an envelope glycoprotein with three main domains (DI-DIII). DII domain mediates membrane fusion and DIII interacts with cellular receptors. |
| HIV gp41 (DDX1306, DDX1304, DDX1305) | gp41 is a unit of the viral envelope glycoprotein complex (gp120/gp41) involved in membrane fusion. |
| Influenza A Hemagglutinin H7N7 (NBP2-89790) | HA is one of two envelope glycoprotein spikes that functions in receptor binding and membrane fusion. |
| MERS-CoV NCoV Spike (NBP2-90336) | Spike is an envelope glycoprotein that forms trimers on the viral surface, binds to cellular receptors, and mediates membrane fusion. |
Branza-Nichita, N., Macovei, A., & Lazar, C. (2012). Caveolae-Dependent Endocytosis in Viral Infection. In Molecular Regulation of Endocytosis. https://doi.org/10.5772/48538
Cruz-Oliveira, C., Freire, J. M., Conceição, T. M., Higa, L. M., Castanho, M. A. R. B., & Da Poian, A. T. (2015). Receptors and routes of dengue virus entry into the host cells. FEMS Microbiology Reviews. https://doi.org/10.1093/femsre/fuu004
Das, A., Hirai-Yuki, A., González-López, O., Rhein, B., Moller-Tank, S., Brouillette, R., … Lemon, S. M. (2017). TIM1 (HAVCR1) is not essential for cellular entry of either quasi-enveloped or naked hepatitis a virions. MBio. https://doi.org/10.1128/mBio.00969-17
Dowd, K. A., & Pierson, T. C. (2011). Antibody-mediated neutralization of flaviviruses: A reductionist view. Virology. https://doi.org/10.1016/j.virol.2010.12.020
Dutta, D., & Donaldson, J. (2012). Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cellular Logistics. https://doi.org/10.4161/cl.23967
Gelderblom, H. R. (1996). Structure and Classification of Viruses. In Medical Microbiology.
Hughey, P. G., Roberts, P. C., Holsinger, L. J., Zebedee, S. L., Lamb, R. A., & Compans, R. W. (1995). Effects of Antibody to the Influenza A Virus M2 Protein on M2 Surface Expression and Virus Assembly. Virology. https://doi.org/10.1006/viro.1995.1498
Ivanov, A. I. (2008). Pharmacological inhibition of endocytic pathways: Is it specific enough to be useful? Methods in Molecular Biology. https://doi.org/10.1007/978-1-59745-178-9_2
Kaushik-Basu, N., Bopda-Waffo, A., Talele, T. T., Basu, A., Costa, P. R. R., Da Silva, A. J. M., … Noël, F. (2008). Identification and characterization of coumestans as novel HCV NS5B polymerase inhibitors. Nucleic Acids Research. https://doi.org/10.1093/nar/gkm1178
Khare, R., Reddy, V. S., Nemerow, G. R., & Barry, M. A. (2012). Identification of Adenovirus Serotype 5 Hexon Regions That Interact with Scavenger Receptors. Journal of Virology. https://doi.org/10.1128/jvi.05760-11
Klasse, P. J. (2014). Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Advances in Biology. https://doi.org/10.1155/2014/157895
Koivusalo, M., Welch, C., Hayashi, H., Scott, C. C., Kim, M., Alexander, T., … Grinstein, S. (2010). Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. Journal of Cell Biology. https://doi.org/10.1083/jcb.200908086
Kondratowicz, A. S., Lennemann, N. J., Sinn, P. L., Davey, R. A., Hunt, C. L., Moller-Tank, S., … Maury, W. (2011). T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire ebolavirus and Lake Victoria marburgvirus. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.1019030108
Laureti, M., Narayanan, D., Rodriguez-Andres, J., Fazakerley, J. K., & Kedzierski, L. (2018). Flavivirus Receptors: Diversity, Identity, and Cell Entry. Frontiers in Immunology. https://doi.org/10.3389/fimmu.2018.02180
Leung, J. Y. S., Ng, M. M. L., & Chu, J. J. H. (2011). Replication of alphaviruses: A review on the entry process of alphaviruses into cells. Advances in Virology. https://doi.org/10.1155/2011/249640
Matsuda, M., Yamanaka, A., Yato, K., Yoshii, K., Watashi, K., Aizaki, H., … Suzuki, R. (2018). High-throughput neutralization assay for multiple flaviviruses based on single-round infectious particles using dengue virus type 1 reporter replicon. Scientific Reports. https://doi.org/10.1038/s41598-018-34865-y
Mazzon, M., & Marsh, M. (2019). Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Research. https://doi.org/10.12688/f1000research.19694.1
Mercer, J., Schelhaas, M., & Helenius, A. (2010). Virus Entry by Endocytosis. Annual Review of Biochemistry. https://doi.org/10.1146/annurev-biochem-060208-104626
Moss, B. (2012). Poxvirus cell entry: How many proteins does it take? Viruses. https://doi.org/10.3390/v4050688
Nayak, B., Kumar, S., DiNapoli, J. M., Paldurai, A., Perez, D. R., Collins, P. L., & Samal, S. K. (2010). Contributions of the Avian Influenza Virus HA, NA, and M2 Surface Proteins to the Induction of Neutralizing Antibodies and Protective Immunity. Journal of Virology. https://doi.org/10.1128/jvi.02135-09
Preta, G., Cronin, J. G., & Sheldon, I. M. (2015). Dynasore - Not just a dynamin inhibitor. Cell Communication and Signaling. https://doi.org/10.1186/s12964-015-0102-1
Rivera-Serrano, E. E., González-López, O., Das, A., & Lemon, S. M. (2019). Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses. ELife. https://doi.org/10.7554/eLife.43983
Thorley, J. A., McKeating, J. A., & Rappoport, J. Z. (2010). Mechanisms of viral entry: Sneaking in the front door. Protoplasma. https://doi.org/10.1007/s00709-010-0152-6
Wang, H., Yang, P., Liu, K., Guo, F., Zhang, Y., Zhang, G., & Jiang, C. (2008). SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Research. https://doi.org/10.1038/cr.2008.15
Yang, N., & Shen, H. M. (2020). Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. International Journal of Biological Sciences. https://doi.org/10.7150/ijbs.45498