Immunohistochemical Staining of HIF-1 alpha in Human Kidney
Staining of HIF-1 alpha in human kidney using NB100-105. Renal tubular epithelium showed moderate membranous, cytoplasmic and nuclear staining, and glomeruli showed faint to moderate nuclear staining.
Flow Cytometry of Myeloma Cells Stained with Alexa Fluor 488 Conjugated HIF-1 alpha Antibody
Analysis using the Alexa Fluor (R) 488 conjugate of NB100-105. Staining of HIF-1 alpha in multiple myeloma cells: H929 cells (0.5 x 10^6) were stained with Alexa Fluor 488(R) conjugated HIF-1 alpha antibody (NB100-105AF488). Image courtesy of Dr. Barbara Muz at Washington University in St. Louis School of Medicine.
Western Blot Analysis of HIF-1 alpha in Nuclear-Enriched and Whole Cell Extracts from Non-Diabetic and T2D Donor Islets
HIF-1-alpha-Antibody-H1alpha67-Western-Blot-NB100-105-img0029.jpg
Immunohistological Staining of HIF-1 alpha in Paraffin Embedded Human Glioblastoma Multiforme
Analysis using the biotin conjugate of NB100-105. Staining of human glioblastoma multiforme.
Staining of HIF-1 alpha in DFOA Treated and non-Treated HeLa Cells
HIF-1 alpha was detected in immersion fixed HeLa human cervical epithelial carcinoma cell line treated with DFOA using 1 ug/mL of mouse anti-HIF-1 alpha monoclonal antibody NB100-105. Cells were stained using a donkey anti-rabbit secondary antibody and counterstained with DAPI (blue).
Detection of HIF-1 alpha by Western Blot in Multiple Cells Lines with Various Treatments
HIF-1-alpha-Antibody-H1alpha67-Western-Blot-NB100-105-img0032.jpg
Staining of HIF-1 alpha in Paraffin Embedded Human Throat Cancer
Analysis using HIF-1 alpha Antibody on human throat cancer tissue. PFA fixed paraffin-embedded sections. Primary antibody dilution 1:100. Image from verified customer review.
Western Blot Analysis of HIF-1 alpha in Human Hepatocytes
Analysis of HIF-1 alpha in human hepatocytes from cancer patient using anti-HIF-1 alpha antibody NB100-105. Western blot image submitted by a verified customer review.
Use of HRP Conjugated HIF-1 alpha Antibody in Western Blotting of COS-7 Nuclear Extracts
Analysis using the HRP conjugate of NB100-105. Detection of 50 ug cobalt chloride induced COS-7 nuclear extracts (NB800-PC26) using NB100-105.
Western Blot of HIF-1 alpha in CD11c+ Cells from C57BL/6 Mice and Densitometric Analysis
HIF-1-alpha-Antibody-H1alpha67-Western-Blot-NB100-105-img0030.jpg
Western Blot Detection of HIF-1 alpha in Nonmalignant and PCa Cell Lines
HIF-1-alpha-Antibody-H1alpha67-Western-Blot-NB100-105-img0031.jpg
Immunohistological Detection of HIF-1 alpha in PDAC and Adjacent Normal Pancreatic Tissue
HIF-1-alpha-Antibody-H1alpha67-Immunohistochemistry-NB100-105-img0033.jpg
Flow Cytometry of H929 Cells Stained with Alexa Fluor 488 Conjugated HIF-1 alpha Antibody
Analysis using the Alexa Fluor(R) 488 conjugate of NB100-105. Staining of HIF-1 alpha in H929 cells using HIF-1 alpha antibody. Flow cytometry image submitted by a verified customer review.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Effects of one- and five-dosage dry needling (1D, 5D) on iNOS, HIF-1 alpha, COX-2, and VEGF expressions in a needling-treated muscle. (a) Representative Western blot images. The quantification of the protein levels for (b) iNOS, (c) HIF-1 alpha, (d) COX-2, and (e) VEGF. Values are expressed as mean ± SD. *Indicates the significant difference (P < 0.05) between the sham groups (s1D and s5D). #Represents the significant difference (P < 0.05) between the 1D and 5D groups. DN: dry needling.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Hypoxia is implicated in the adaptive response after short-term bevacizumab treatment. Expression of HIF-1 alpha in pulmonary tumor nodules of the four groups. (A) A representative western blot is shown. beta-actin was used as a loading control. (B) While most tumors showed little expression of HIF-1 alpha protein in PBS and cisplatin groups, mice that received bevacizumab and bevacizumab + cisplatin therapy showed a markedly increased level of HIF-1 alpha expression.. *P < 0.05, **P < 0.01.
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Effects of F2 on tumor-related biomarkers in vivoImmunohistochemical analysis of tumor-related proteins (HIF-1 alpha, VEGF, MMP-2, CD31 and KI67) in tumor sections from U87 xenograft mice treated with F2 (mg/kg i.g.). Red arrows indicate the related protein detected in the tissue samples. Values are means ± SEM of three experiments. * P<0.05 compared with DDW group.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
AAM inhibits hypoxia-induced VM formation involving HIF-1 alpha and MMP2. The HCT-116 (A) and LoVo (B) cells were treated with different concentrations of AAM under hypoxia and normoxia; the protein expression of HIF-1 alpha and MMP2 was detected by western blotting. (C, D) The mRNA expression of HIF-1 alpha and MMP2 was measured by RT-PCR (*P < 0.05, **P < 0.01, ***P < 0.001).
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemical analysis of STC1 expression in esophageal cancer. (A) Normal squamous epithelium was negative for STC1 expression. (B) Normal gastric glands showed positive staining for STC1, which was used as a positive control. (C) Positive staining for STC1 in esophageal squamous cell carcinoma (ESCC). (D) Negative staining for STC1 in ESCC. (E) Positive staining for HIF-1 alpha in ESCC. (F) Positive staining for p53 in ESCC. Magnification, ×100.
Simple Western: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Simple Western: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Effect of hypoxia on human airway epithelial cells (A549).A549 cells were subjected to hypoxia for 6, 24, and 48 hours. Cell lysates were collected and analyzed for the expression levels of various hypoxic markers by qPCR (n = 4). (A-C) The expression levels of HIF-1 alpha (A), GLUT-1 (B), and VEGFA (C) were measured. (D-G) Additionally, whole-lung lysate was used for Western blots to determine the protein expression of HIF-1 alpha, VEGFA, and FIH-1. After analyzing the data, we used the unpaired t test with Welch's correction to determine its statistical significance. *P < 0.05, ***P < 0.001, ****P < 0.0001. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/37737265 ), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
The HIF-1 alpha-BNIP3 pathway was activated during hypoxia. (a) Analysis of mitophagy-related protein expression in Neuro-2a cells by immunoblotting. Neuro-2a cells were exposed to hypoxia and/or high glucose for 48 h. Total cellular extracts were analysed by immunoblotting with antibodies against HIF-1 alpha, BNIP3, PINK1 & TOMM20. (b) The ImageJ densitometric analysis is shown. “C” represented control conditions (25 mM glucose), “G” represented high glucose conditions (75 mM glucose). The results are expressed as the mean ± SEM from three independent experiments. Two-way ANOVA, **p < 0.01, *p < 0.05, Hypoxia vs Normoxia. Full-length blots are presented in Supplementary Fig. S2. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-018-20162-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Inhibition of tumor growth in vivo by various treatments. (a) Macroscopic view of mouse tumors at the end of the study. (b) Tumor growth curves from day 0 to 40, with tumor sizes measured every 5 days, in various treatment groups as indicated. (c) Wet weight of tumors measured at the end of the study. (d) Protein expression of HIF-1 alpha examined by western blot in PC-3 xenografts exposed to various treatments. Data were presented as mean ± SD (n = 6). *p < 0.05 versus control group; #p < 0.05 versus si-HIF-1 alpha or DDP group. The original blots are presented in Supplementary Figure 6. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-07973-4), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - AAM inhibits hypoxia-induced VM formation involving HIF-1 alpha & MMP2. The HCT-116 (A) & LoVo (B) cells were treated with different concentrations of AAM under hypoxia & normoxia; the protein expression of HIF-1 alpha & MMP2 was detected by western blotting. (C, D) The mRNA expression of HIF-1 alpha & MMP2 was measured by RT-PCR (*P < 0.05, **P < 0.01, ***P < 0.001). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32499699), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Muscle repair following eccentric contraction–induced injury is concomitant with dynamic alterations of HIF2A & HIF1A expression in SCs.(A–C) Representative images of EDL myofibers from injured muscles at various time points (n >50 myofibers from 3 mice/group/time point) & stained for Pax7, DAPI, & EdU (A), HIF2A (B), or HIF1A (C). Scale bars: 20 μm. Arrowheads indicate SCs. (D) Number of Pax7+ SCs per myofiber at various time points. (E) Percentage of EdU+ SCs at various time points. (F) Percentage of HIF2A+ SCs at various time points. (G) Percentage of HIF1A+ SCs at various time points. Data represent the mean ± SEM. Image collected & cropped by CiteAb from the following publication (https://www.jci.org/articles/view/96208), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - mTORC2 not mTORC1 regulates HIF‐2 alpha, GOT1 in PDAC under prolonged hypoxia. (A & B) HIF‐2 alpha mRNA & protein levels determined by qRT‐PCR (mRNA) & Western blot (protein) after treatment of Panc‐1 & Capan‐2 cells with mTORC1 inhibitor rapamycin for 48 hrs at 3% or 1% O2. beta‐Actin was used as loading control. (C & D) Panc‐1 & Capan‐2 cells were treated with mTORC1/mTORC2 inhibitor PP242 & cultured for 48 hrs at 20%, 3% or 1% O2, & HIF‐2 alpha, GOT1 mRNA & protein levels were determined by qRT‐PCR (mRNA) & Western blot (protein). beta‐Actin was used as loading control. Data are presented as mean ± S.D. from three independent experiments. *P < 0.05, **P < 0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28544376), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - mTORC2 not mTORC1 regulates HIF‐2 alpha, GOT1 in PDAC under prolonged hypoxia. (A & B) HIF‐2 alpha mRNA & protein levels determined by qRT‐PCR (mRNA) & Western blot (protein) after treatment of Panc‐1 & Capan‐2 cells with mTORC1 inhibitor rapamycin for 48 hrs at 3% or 1% O2. beta‐Actin was used as loading control. (C & D) Panc‐1 & Capan‐2 cells were treated with mTORC1/mTORC2 inhibitor PP242 & cultured for 48 hrs at 20%, 3% or 1% O2, & HIF‐2 alpha, GOT1 mRNA & protein levels were determined by qRT‐PCR (mRNA) & Western blot (protein). beta‐Actin was used as loading control. Data are presented as mean ± S.D. from three independent experiments. *P < 0.05, **P < 0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28544376), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Murine tumor cells express connexin 43 (Cx43) & hypoxic-induced factor-1 alpha (HIF-1 alpha). The expression of Cx43 & HIF-1 alpha was measured by Western blot analysis. beta-actin expression served as loading controls & total protein. Inserted values indicated relative protein expression in comparison with beta-actin. Image collected & cropped by CiteAb from the following publication (http://www.mdpi.com/1422-0067/16/1/439), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Representative microscopy images of staining for hypoxia markers in prostate tissues (MO, 400×). A) HIF-1 alpha - notice the granular cytoplasmic immunoreactivity of the malignant epithelial cells. In this case, more than 50% of the glands stained. B) LOX - strong & diffuse nuclear immunoreactivity of the epithelial cells. C) CAIX - note a focal apical cytoplasmic immunoreactivity in epithelial cells. D) VEGFR2 - moderate nuclear & weak cytoplasmic expression of the epithelial cells Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28143503), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Effect of renal medullary transfection of PHD2 plasmids on the levels of PHD2 & HIF-1 alpha in the renal medulla. (A) PHD2 mRNA levels. (B) Representative ECL gel documents of Western blot analyses depicting the protein levels of HIF-1 alpha. (C) Summarized intensities of the HIF-1 alpha blots (normalized to LS). *P < 0.05 versus others (n = 7–8). LS = low salt, HS = high salt, Ctrl = control vectors, PHD2 = PHD2 expression vectors, CoCl2 = sample from cells treated with CoCl2 as positive control. Renal medullary tissues were obtained at the end of blood pressure recording after 3 week high salt challenge. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/22686466), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Immunohistochemical analysis of STC1 expression in esophageal cancer. (A) Normal squamous epithelium was negative for STC1 expression. (B) Normal gastric glands showed positive staining for STC1, which was used as a positive control. (C) Positive staining for STC1 in esophageal squamous cell carcinoma (ESCC). (D) Negative staining for STC1 in ESCC. (E) Positive staining for HIF-1 alpha in ESCC. (F) Positive staining for p53 in ESCC. Magnification, ×100. Image collected & cropped by CiteAb from the following publication (https://www.spandidos-publications.com/10.3892/or.2011.1607), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1alpha & CAIX expression in PDAC & adjacent normal pancreatic tissue. Adjacent normal exocrine pancreas with weak nuclear & cytoplasmic HIF-1alpha staining (a). Adjacent normal pancreatic tissue with weak membranous CAIX staining in the ducts & absent staining in exocrine pancreatic tissue (b). PDAC with weak (intensity 1) nuclear & cytoplasmic HIF-1alpha staining (c). PDAC with strong (intensity 3) nuclear & moderate (intensity 2) cytoplasmic HIF-1alpha staining (d). PDAC with weak to moderate (intensity 1–2) membranous CAIX staining (e). PDAC with strong (intensity 3) membranous CAIX staining (f) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29973215), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha staining within injured muscle area (representative figure).The animal was ventilated with 100% O2. 450 minutes after trauma, the Musculus gastrocnemius of the traumatized right hind limb was harvested & section of the muscle specimen was analyzed for HIF-1 alpha expression within the injured area. No significant staining for HIF-1 alpha of the muscle cells. Scale bar: 150 µm. Inlet: Positive staining for HIF-1 alpha expression by myeloid cells (brown), invading to the traumatized muscle. Scale bar: 10 µm. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0111151), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF1 alpha is activated in VSMCs during Ang II-induced vascular remodeling.WT mice were infused with saline or 1000 ng/kg/min Ang II for 28 days. a Immunofluorescence analysis of representative cross-sections of mice aortas for HIF1 alpha (red) & alpha-SMA (green), nuclei was stained with DAPI. VSMCs were isolated form WT mice & treated with 1 μM Ang II for 24 h. bHif1a mRNA was measured by qPCR analysis. c HIF1 alpha protein was detected by western blot. **P < 0.01, ***P < 0.001, n = 3 per group, statistical significance was determined by the unpaired t-test Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31320613), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Hypoxia is implicated in the adaptive response after short-term bevacizumab treatment. Expression of HIF-1 alpha in pulmonary tumor nodules of the four groups. (A) A representative western blot is shown. beta-actin was used as a loading control. (B) While most tumors showed little expression of HIF-1 alpha protein in PBS & cisplatin groups, mice that received bevacizumab & bevacizumab + cisplatin therapy showed a markedly increased level of HIF-1 alpha expression.. *P < 0.05, **P < 0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/22357313), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1alpha & CAIX expression in PDAC & adjacent normal pancreatic tissue. Adjacent normal exocrine pancreas with weak nuclear & cytoplasmic HIF-1alpha staining (a). Adjacent normal pancreatic tissue with weak membranous CAIX staining in the ducts & absent staining in exocrine pancreatic tissue (b). PDAC with weak (intensity 1) nuclear & cytoplasmic HIF-1alpha staining (c). PDAC with strong (intensity 3) nuclear & moderate (intensity 2) cytoplasmic HIF-1alpha staining (d). PDAC with weak to moderate (intensity 1–2) membranous CAIX staining (e). PDAC with strong (intensity 3) membranous CAIX staining (f) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29973215), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - DC101, evofosfamide, & low dose doxorubicin multimodal treatment of KP sarcomasA. Tumor volume of KP sarcomas. Groups were treated by intraperitoneal injection of control IgG 20 mg/kg 3 times per week, DC101 20 mg/kg 3 times per week, evofosfamide (Evo) 50 mg/kg 5 times per week, and/or low dose doxorubicin (Dox) 1 mg/kg 3 times per week. Photos B. & graphs C. of PCNA immunohistochemistry for proliferation, TUNEL immunofluorescence for apoptosis, CD31 immunohistochemistry for microvessel density, CD31 & TUNEL immunofluorescence for endothelial cell (EC)-specific apoptosis, & nuclear HIF-1 alpha immunohistochemistry for HIF-1 alpha activity. Bars represent standard deviation. **p < 0.05 compared to all other groups. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.10212), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Overexpression of HIF alpha proteins increases IL-1 alpha transcription in A549 & HeLa cells but not in WI-38 fibroblasts.(A) Western blot using anti-HIF-1 alpha or anti-HIF-2 alpha antibodies of nuclear fractions obtained from HEK-T293 cells transfected with the annotated vectors. WI-38 (B), A549 (C), or HeLa (D) cells were transfected with either control vector or with HIF-1 alpha (P402A/P564A) & HIF-2 alpha (P405A/P531A), & 24 h later were analyzed for IL-1 alpha & VEGF RQ of mRNA levels by real-time PCR. Graphs represent mean ± SEM of four independent experiments for each cell type. P values of HIF alpha-transfected cells vs. control (Ct) transfected are annotated in the graphs. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23049530), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Effects of F2 on tumor-related biomarkers in vivoImmunohistochemical analysis of tumor-related proteins (HIF-1 alpha, VEGF, MMP-2, CD31 & KI67) in tumor sections from U87 xenograft mice treated with F2 (mg/kg i.g.). Red arrows indicate the related protein detected in the tissue samples. Values are means ± SEM of three experiments. * P<0.05 compared with DDW group. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.19654), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Immunohistochemical labeling of normal brain & glioma tissueRepresentative images of normal temporal lobe, anaplastic astrocytoma grade II & grade III (AAII & AAIII), & three GBMs with low, medium, & high labeling of (A) MAOB, (B) HiF-1 alpha, & (C) GFAP are shown, along with (D) Sp1 & e) Sp3. The antibodies were detected with DAB & the nuclei were labeled with hematoxylin. Each image in a column was taken from a single slide, where each tissue slice was treated identically & imaged using the same microscope setting & with no electronic image manipulation. Arrows indicate nuclear HiF-1 alpha. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.6582), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Expression of PACAP, VIP, VIPRs, HIF-1 alpha, HIF-2 alpha, & EGFR in glioblastoma multiforme (GBM). (A) Representative immunoblot of PACAP & VIP precursor peptides & PAC1R, VPAC1R, & VPAC2R expression on frozen glioblastoma sample. (B) Representative immunoblot & photomicrographs of signals detected by antibodies direct against HIF-1 alpha, HIF-2 alpha, & EGFR in a frozen glioblastoma sample. Image collected & cropped by CiteAb from the following publication (http://journal.frontiersin.org/Article/10.3389/fphar.2016.00139/abstract), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Regulation of cartilage degradation by HIF-1 alpha. (A) Amount of glycosaminoglycans released into the medium determined by the dimethylmethylene blue assay during 3 days culture of Hif1afl/fl (Cntl) & Col2a1-CreERT2;Hif1afl/fl (KO) femoral heads treated with or without 10 ng/mL IL-1 beta under the hypoxic condition (3% O2). Tamoxifen induction was performed three days before sacrifice. Bars show the mean ± SD of three samples per group. *P < 0.05. (B) Immunofluorescence of Mmp13, Hif2a, & Hif1a in the Cntl & KO femoral head cartilage after 3-day culture with or without 10 ng/mL IL-1 beta. Scale bars, 50 µm. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32214220), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF1 alpha/PFKFB3 is upregulated in beta-cells of HIP rats & humans with type 2 diabetes. Representative immunofluorescence images of islets from a WT & HIP rats & b non-diabetic (ND) & T2D patients stained for PFKFB3 (red), insulin (green), & nuclei (blue). c Frequency of PFKFB3 positive beta-cells in HIP versus WT rats (left panel) & T2D versus ND patients (right panel). d Representative Western blot of PFKFB3 & HIF1 alpha levels in nuclear-enriched- & whole cell extracts from non-diabetic (ND) & T2D donor islets. Data are presented as mean ± SEM, n = 3 independent biological samples for each group. Statistical significance was analyzed by Student t-test (*p < 0.05, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31213603), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha & HIF-2 alpha expression is differentially modulated by Hsp90 inhibition in CCRCC cells. UMRC2 & 786-O cells were treated for 20 h with the Hsp90 inhibitors 17-AAG (1 μM), EC154 (100 nM), the HDAC inhibitor LBH589 (100 nM), or DMSO vehicle control. Nuclear protein was analyzed for HIF-1 alpha & HIF-2 alpha & cytosolic protein was analyzed for RACK1 by SDS-PAGE & Western blot. Topoisomerase II & GAPDH were used as nuclear & cytosolic loading controls, respectively. Bands corresponding to HIF-1 alpha & HIF-2 alpha Western blots were quantified, normalized to TOPOII, & are expressed as a percentage of control samples. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/22172030), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Comparison of MAOB expression, HiF-1 alpha expression, & peroxide generation in response to a MAOB substrate in normal human astrocytes & glioma cellsHiF-1 alpha (i, red), MAOB (ii, green), & superimposed HiF-1 alpha/MAOB signals with blue DAPI labeling (iii) of NHAs (A) & GBM cells (B). (C) Relative expression of HiF-1 alpha (white bars) & MAOB (black bars) in NHAs & in three GBM low-passage cultures, n = 6, mean ± SD. (D) MAOB inhibited (gray bars), endogenous (white bars) & 4-FBA-stimulated (black bars) reactive oxygen species (hydrogen peroxide) generation, shown as DCF per cell, in NHA & the three GBM cultures, n = 16, mean ± SD. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.6582), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF1 alpha is activated in VSMCs during Ang II-induced vascular remodeling.WT mice were infused with saline or 1000 ng/kg/min Ang II for 28 days. a Immunofluorescence analysis of representative cross-sections of mice aortas for HIF1 alpha (red) & alpha-SMA (green), nuclei was stained with DAPI. VSMCs were isolated form WT mice & treated with 1 μM Ang II for 24 h. bHif1a mRNA was measured by qPCR analysis. c HIF1 alpha protein was detected by western blot. **P < 0.01, ***P < 0.001, n = 3 per group, statistical significance was determined by the unpaired t-test Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31320613), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Effects of one- & five-dosage dry needling (1D, 5D) on iNOS, HIF-1 alpha, COX-2, & VEGF expressions in a needling-treated muscle. (a) Representative Western blot images. The quantification of the protein levels for (b) iNOS, (c) HIF-1 alpha, (d) COX-2, & (e) VEGF. Values are expressed as mean ± SD. *Indicates the significant difference (P < 0.05) between the sham groups (s1D & s5D). #Represents the significant difference (P < 0.05) between the 1D & 5D groups. DN: dry needling. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23346198), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Effect of ENMD-1198 on tumor vascularization & tumor cell proliferation. A) Immunohistochemical analysis of tumor vascularization (CD31-positive vessels). Densitometric analysis of images from CD31-stained tissue sections from all tumors showed that vessel area was significantly reduced in ENMD-1198-treated tumors (*P < 0.01). B) ENMD-1198 significantly decreased the number of proliferating (BrdUrd-positive) tumor cells in tissue sections (*P < 0.01). For both vessel area & cell proliferation, representative images are illustrated. Results shown are the mean ± SEM. C) Western blot analysis for HIF-1 alpha expression in HCC tumors showed a substantial reduction of HIF-1 alpha in ENMD-1198 treated tumors. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/18651980), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Enhanced expression of glycolytic genes in livers of DEN-Wwox deltaHep mice.a mRNA expression levels of HIF1 alpha glycolytic target genes in DEN-treated control & Wwox deltaHep (cKO) mice at the age of 1 a, 3 b, 6 c & 10 d months (n = 3 for each group). e Immunohistochemical staining of HIF1 alpha, PKM2, & GLUT1 in 6 & 10 months of DEN-treated Wwox deltaHep & control liver tissues. Images were taken at × 40 magnification (Bar = 100μm). f ChIP experiment with HIF1 alpha antibody on DEN-treated control & Wwox deltaHep (cKO) mice followed by qRT-PCR analysis of HIF1 alpha target genes. * P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41419-018-0510-4), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1alpha & CAIX expression in PDAC & adjacent normal pancreatic tissue. Adjacent normal exocrine pancreas with weak nuclear & cytoplasmic HIF-1alpha staining (a). Adjacent normal pancreatic tissue with weak membranous CAIX staining in the ducts & absent staining in exocrine pancreatic tissue (b). PDAC with weak (intensity 1) nuclear & cytoplasmic HIF-1alpha staining (c). PDAC with strong (intensity 3) nuclear & moderate (intensity 2) cytoplasmic HIF-1alpha staining (d). PDAC with weak to moderate (intensity 1–2) membranous CAIX staining (e). PDAC with strong (intensity 3) membranous CAIX staining (f) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29973215), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Chromatin Immunoprecipitation: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Chromatin Immunoprecipitation: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Involvement of CBP in BNIP3 expression regulated by HIF-1 alpha & FOXO3 under hypoxia. (A) UCB-hMSCs were incubated with hypoxia condition for 24 h. Co-immunoprecipitation of HIF-1 alpha & FOXO3 with IgG & CBP were shown in left panel. IgG was used as a negative control. The total protein expressions of HIF-1 alpha, FOXO3, CBP & beta-actin in lysate were shown in right panel. n = 3. (B) CBP (20 μM) was pretreated to UCB-hMSCs, & cells were incubated with hypoxia for 24 h. The BNIP3 mRNA expression level was analyzed by qPCR. n = 6. (C) BNIP3 & beta-actin protein expressions were analyzed by western blot. Data represent mean ± S.E. n = 4. (D, E) Sample DNA was immuno-precipitated with RNA polymerase, IgG, HIF-1 alpha & FOXO3 specific antibodies. CHIP (top panel) & lysate (bottom panel) samples were amplified with the primers of GAPDH & BNIP3 promoters. Quantitative CHIP data was analyzed by qPCR, & shown in the right panel. n = 4. Western blot data were normalized by beta-actin, & qPCR data were normalized by ACTB mRNA expression level. Quantitative data are presented as a mean ± S.E.M. All blot images are representative. *p < 0.05 versus control, #p < 0.05 versus hypoxia. Image collected & cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213231717303804), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Lack of HIF-P4H-1 increases the amount of p53. (a) Western blot analysis of p53 in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h. (b,c) Western blot analysis of p53 in Hif-p4h-1 & scrambled (Scrm) siRNA transfected HEK293 cells (b) & wt MEFs treated with 50 µM FG4497 for 24 h (c). (d) qPCR analysis of p53 mRNA in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h. (e) Analysis of p53 protein turnover rate. Hif-p4h-1−/− & wt MEFs were treated with 200 μg/ml of cycloheximide for the indicated time points & whole cell lysates were blotted for p53. (f) Western blot analysis of MDM2 in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h. (g) Western blot analysis of p53 & MDM2 in wt MEFs treated with 10 μM nutlin-3a for 24 h with or without overexpression of human HIF-P4H-1 (OE). (h,i) Western blot analysis of HIF1 alpha in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h (h) & in scrambled & Hif-p4h-1 siRNA transfected HEK293 cells (i). (j) Western blot analysis of ubiquitination of p53 in Hif-p4h-1 & scrambled siRNA transfected HEK293 cells. The cells were transfected with HA-ubiquitin & endogenous p53 was immunoprecipitated followed by Western blotting with anti-HA & anti-P53 antibodies. (k) Western blot analysis of p53 in Hif-p4h-1−/− & wt MEFs were treated with or without 10 μM MG132 for 5 h. Data are presented as representative Western blots & as mean ± s.d., n = at least 3 individual MEF isolates or experiments. *P < 0.05, **P < 0.01 & ***P < 0.001, two-tailed Student’s t-test. Unprocessed original scans of blots are shown in Supplementary Fig. 5. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-17376-0), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha-mediated repression of ENT1 expression in Caki-1 cells under hypoxia. Comparison of HIF-1 alpha & HIF-2 alpha protein expression in human matched renal normal-tumor samples (n=11) (A) & 3 RCC cell lines (786-O, 769-P, Caki-1) under normoxia or hypoxia for 6 h (B). (C) Schematic diagram of two hypoxia response elements (HRE) in the promoter of ENT1. (D) CHIP-qPCR analysis of HIF-1 alpha at the promoter of ENT1 in Caki-1 cells, exposed under normoxia or hypoxia for 48 h. Two different HIF-1 alpha antibodies (610958, BD Biosciences; NB100-105, Novus) were used to pull down HIF-1 alpha. The protein expression level of HIF-1 alpha (E) & mRNA expression level of SLC29A1 (F) in Caki-1 under normoxia or hypoxia & HIF-1 alpha knockout-Caki-1 cells. (G) The expression level of ARNT1 (HIF-1 beta) was detected in Caki-1, transfected with ARNT1 specific siRNAs or NC. Student's t test (two-tailed) was used. (H, I) The expression level of ENT1 in Caki-1, transfected with ARNT1 specific siRNAs or NC, under normoxia or hypoxia. Oneway-ANOVA analysis was used. The results are expressed as mean ± SEM (n=3). ***P<0.001. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32206108), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - L. donovani infection induces HIF-1 alpha expression in CD11chi splenic DCs in an IRF-5 dependent manner.Mice were infected with 2x107 amastigotes intravenously. (A) Real-time PCR analysis of HIF-1 alpha mRNA expression in CD11c+ cells purified from C57BL/6 mice at various time points after infection. (B) Immunoblot analysis of HIF-1 alpha expression in CD11c+ cells from C57BL/6 mice (upper panel) & densitometric analysis normalized to beta-actin expression & expressed as fold increase to results obtained with naïve mice (lower panel). (C) Real-time PCR analysis of HIF-1 alpha expression in sorted CD11c+ cells from Irf-5flox/floxCre- & Irf-5flox/floxCMV-Cre+. (D) Immunoblot analysis of Hif-1 alpha expression in CD11c+ cells population of Irf5flox/floxCre- (left upper panel) & Irf-5flox/floxCMV-Cre+ (right upper panel), & densitometric analysis normalized to beta-actin expression & expressed as fold increase to results obtained with naïve mice (lower panels). (E) Real-time PCR analysis of Hif-1 alpha expression in CD11c- splenocytes from Irf-5flox/floxCre- & Irf-5flox/floxCMV-Cre+. (F) Real-time PCR analysis of HIF-1 alpha mRNA expression in BMDC from Irf-5flox/floxCMV-Cre+ & Cre- mice. All data represent mean ± SEM combined from 3 independent experiments. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.ppat.1004938), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Optical redox ratio (ORR) demonstrates reduced glycolytic phenotype following 24 hours of YC-1 treatment. ORR of A549 & A549RR cells before, 12 & 24 hours following 50 μM YC-1 treatment to inhibit HIF-1 (a,b). Immunoblotting for HIF-1 alpha (c) & PDK-1 (d) before & 24 hours following YC-1 treatment to verify in of HIF-1 alpha. Representative immunoblots were horizontally cropped at the molecular weight indicated (e). Uncropped exposures can be seen in Supplemental Fig. 1. Experiments were performed in triplicate across 3 independent experiments. *p < 0.05 compared to A549 0 hr control, †p < 0.05 compared to A549RR 0hrs. Scale bar in images represents 50 μm. ME – Main effect of cell type & time; however, no significant interactions were found between the two main effects. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-018-27262-y), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - A beta facilitates HIF1 alpha synthesis & autophagy inhibition via mTOR activation. (A) SK-N-MC cells exposed to A beta (5 μM) for 0–48 h. HIF1 alpha & beta-actin expression analyzed by WB. n = 3. (B) Cells pretreated w/ NAC (1 mM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expression analyzed by WB. n = 3. (C,E) Cells incubated w/ rapamycin (10 nM) for 30 min prior to A beta treatment for 24 h. Phosphorylation of 4EBP1 (Thr 37/46) & 4EBP1, phosphorylation of p70S6K1 (Thr 389), HIF1 alpha & beta-actin analyzed by WB. n = 6. (D) Protein samples immunoprecipitated by eukaryotic translation initiation factor 4E (eIF4E) antibody-conjugated protein A/G agarose beads. Samples blotted w/ 4EBP1 & eIF4E-specific antibodies. n = 3. (F) Cells exposed to PF4708671 (10 μM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expression detected by WB. n = 6. (G) Cells exposed to cycloheximide (4 μM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expressions detected by WB. n = 6. (H) Cells pretreated w/ rapamycin (10 nM) for 30 min, incubated w/ A beta for 24 h & analyzed by WB w/ LC3, p62 & beta-actin specific antibodies. n = 3–6. (I) LC3 puncta visualized by confocal microscopy. Presented results merged images. Green & red fluorescents indicate LC3 & PI respectively. Scale bars, 50 μm (magnification × 600). (J) Cells pretreated w/ trehalose (10 μM) for 30 min prior to A beta treatment for 24 h. Cytotoxicity measured by MTT assay at an absorbance of 545 nm using a microplate reader. Data present the mean ± SE. n = 6. (K) Cell viability measured by trypan blue exclusion assay. Data presented as a mean ± SE. n = 6. Each blot image presented as representative image. *p < 0.05 vs. control, #p < 0.05 vs. A beta treatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28790888), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - FDFT1 inhibits glycolysis through suppressing AKT-mTOR-HIF1 alpha pathway in CRC.aFDFT1 overexpression reduced glucose uptake in CT26 cells. bFDFT1 overexpression decreased lactate production via glycolysis in CT26 cells. c ECAR reduced when FDFT1 overexpressed in CT26 cells. d OCR increased when FDFT1 overexpressed in CT26 cells. eFDFT1 knockdown increased glucose uptake in CT26 cells. fFDFT1 knockdown increased lactate production via glycolysis in CT26 cells. g ECAR increased when FDFT1 knocked down in CT26 cells. h OCR decreased when FDFT1 knocked down in CT26 cells. i, mFDFT1 overexpression inhibited protein & mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, & LDHA, in CT26 cells. j, mFDFT1 knockdown increased protein & mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, & LDHA, in CT26 cells. k, nFDFT1 overexpression decreased protein & mRNA expression of AKT, mTOR, & HIF1 alpha. l, nFDFT1 knockdown increased protein & mRNA expression of AKT, mTOR, & HIF1 alpha. o Photograph of dissected tumors (the first line: normal diet, second line: FMD + glucose, third line: FMD, n = 5). p The tumor volumes measured every 2 days after 13th day. The FMD + glucose group can reverse tumor growth inhibition induced by FMD (n = 5; ns: P = 0.1838; P = 0.0001). q The protein level of FDFT1 & mTOR in dissected tumor samples from normal diet group, FMD group & FMD + glucose group measured by western blotting. r The glucose level in these three groups. Error bars, mean ± SD, data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared w/ control group (or normal diet group). #P < 0.05, ##P < 0.01. Image collected & cropped by CiteAb from following publication (https://pubmed.ncbi.nlm.nih.gov/32313017), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - L. donovani infection induces HIF-1 alpha expression in CD11chi splenic DCs in an IRF-5 dependent manner.Mice were infected with 2x107 amastigotes intravenously. (A) Real-time PCR analysis of HIF-1 alpha mRNA expression in CD11c+ cells purified from C57BL/6 mice at various time points after infection. (B) Immunoblot analysis of HIF-1 alpha expression in CD11c+ cells from C57BL/6 mice (upper panel) & densitometric analysis normalized to beta-actin expression & expressed as fold increase to results obtained with naïve mice (lower panel). (C) Real-time PCR analysis of HIF-1 alpha expression in sorted CD11c+ cells from Irf-5flox/floxCre- & Irf-5flox/floxCMV-Cre+. (D) Immunoblot analysis of Hif-1 alpha expression in CD11c+ cells population of Irf5flox/floxCre- (left upper panel) & Irf-5flox/floxCMV-Cre+ (right upper panel), & densitometric analysis normalized to beta-actin expression & expressed as fold increase to results obtained with naïve mice (lower panels). (E) Real-time PCR analysis of Hif-1 alpha expression in CD11c- splenocytes from Irf-5flox/floxCre- & Irf-5flox/floxCMV-Cre+. (F) Real-time PCR analysis of HIF-1 alpha mRNA expression in BMDC from Irf-5flox/floxCMV-Cre+ & Cre- mice. All data represent mean ± SEM combined from 3 independent experiments. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.ppat.1004938), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - WNT11 is induced by hypoxia or hypoxic mimetics in different cell types.(A) Increased Wnt11 mRNA in EMSC adipocytes (Day 12) after hypoxia-mimetic treatments. EMSC adipocytes were treated with CoCl2 (0.1 mM), DFO (0.1 mM) or DMOG (0.1 mM) for 24 hrs. Values were normalized to Tbp mRNA & are expressed relative to control (n = 3). (B,C) Increased Wnt11 mRNA by hypoxia in EMSC preadipocytes & adipocytes (Day 0–12 after differentiation) (B), & C2C12 myoblast & myocyte (Day 0 & 8 after differentiation) (C). Wnt11 mRNA was assessed by quantitative PCR in cells exposed to air (21% O2) or hypoxia (1% O2) for 24 hrs. (n = 4). Values were normalized to Tbp mRNA & are expressed relative to 21% O2 samples (left panel). (D) Immunoblot analyses of HeLa cells under normal air or hypoxia for 24 hrs. (E,F) Induction of Wnt11 by increasing concentrations of DMOG in MDA-MB-231 cells (E) & 4T1 cells (F). (G) EMSCs treated with 0.1 mM DMOG for the indicated times. Wnt11 & Vegf mRNA expression was measured by qPCR & normalized to Tbp mRNA (n = 4). (H) WNT11 protein levels after DMOG treatment normalized to alpha-Tubulin (upper panel; n = 4). Representative immunoblots of EMSCs treated with 0.1 mM DMOG for the indicated times (Lower panel). (I) Protein expression in MDA-MB-231 cells treated with 0.1 mM DMOG. (J) Induction of Wnt11 promoter activity by hypoxia or hypoxia mimetics. pGL3-Wnt11 promoter plasmid was transfected into C2C12 cells. Cells were incubated with DMOG (left panel, n = 4) or under 21% O2 or 1% O2 (right panel, n = 8) for 24 hrs. For panels (A–C,G,H,J), values are mean ± s.e.m. *p < 0.05, **p < 0.01. For panels of immunoblotting, laminin, alpha-tubulin, & ERK were used as loading controls, WNT11 normalized to alpha-Tubulin was shown. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/srep21520), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Ccl7 is a HIF1 alpha direct target gene.a qPCR analysis of Ccl7 mRNA expression in Hif1afl/fl & Hif1a deltaSMC VSMCs treated with vehicle, CoCl2, normoxia, or hypoxia (2% O2) for 6, 12, & 24 h. *P < 0.05, **P < 0.01, ***P < 0.001, n = 6 (independent experiments) per group. VSMCs isolated from Hif1afl/fl mice & Hif1a deltaSMC mice were infected with oxygen-stable HIF1 alpha-expressing lentivirus, & then treated with Ang II for 24 h, bCcl7 mRNA was measured by qPCR & c CCL7 protein was detected by ELISA. d qPCR analysis of Ccl7 mRNA expression in vehicle or Ang II-treated Hif2afl/fl & Hif2a deltaSMC VSMCs. e Schematic diagram of the mouse Ccl7 promoter illustrating the HREs in the regulatory region; the upstream regions were numbered in relation to the transcription initiation site. f Luciferase-reporter constructs under the control of the mouse Ccl7 promoter. HEK293T human embryonic kidney cells transiently transfected with the luciferase construct, & cotransfected with empty vector or HIF1a expression plasmids. Standard dual-luciferase assays were performed. EV, empty vector. **P < 0.01, n = 3. g, h ChIP assays of vehicle or Ang II-treated wild-type VSMCs using HIF1 alpha or HIF2 alpha antibodies. Data were normalized to input. *P < 0.05, **P < 0.01, n = 6 per group. i, j ChIP assays of vehicle or Ang II-treated Hif1afl/fl & Hif1a deltaSMC VSMCs using HIF1 alpha antibody. Data were normalized to input. *P < 0.05, **P < 0.01, n = 6 per group. Statistical significance was determined by one-way ANOVA test followed by the unpaired t-test Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31320613), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Hypoxic/necrotic core in GemOE tumors enhances Ac-HMGB1 secretion(A–C) Adjacent sections from a GemOE tumor IHC stained for geminin (A), c-Abl (B) & HMGB1 (C). (D) The level of circulating HMGB1 measured using specific ELISA assay performed on serum isolated from samples collected 7 weeks after mice were injected in mammary fat pads with naïve HME cells (n = 10, no tumors developed) or GemOE cells (n = 30, tumor-bearing, p = 0.00042). Two different sets (E, G, I & K) & (F, H, J & L) of adjacent sections from GemOE orthotopic mammary tumors stained with H & E (E & F), or IHC stained for geminin (G & H), HMGB1 (I & J) as well as HIF-1 alpha (K) or hypoxyprobe (L). N denotes necrosis within these tumors that are shown adjacent to the hypoxic cells as indicated by high HIF-1 alpha or hypoxyprobe staining. These cells are also expressing cytoplasmic HMGB1. (M) The levels of HMGB1 detected using specific ELISA assay released from iGem9, iG197, iG240 or iG257 cells grown under normoxic (N) or hypoxic (H) conditions. Experiments were done in triplicates 3 different times, **represents p < 0.001. (N) The level of acetylated HMGB1 passively diffused from naïve HME, iG197, iG240 or iG257 cells after repeated freeze & thaw cycles. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26989079), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - A beta induces the expression of cell cycle regulatory proteins. (A) SK-N-MC cells were exposed to A beta (5 μM) for 24 h. The mRNA expression levels of CDK5, P35 & P39 were analyzed by real-time PCR. The mRNA expression level was normalized by beta-actin mRNA expression level. Data represent the mean ± SE. n = 4. (B)hif1 alpha specific- & non-targeting (NT) siRNA were transfected to the cells for 24 h prior to A beta treatment. Cyclin D1, CDK4, cyclin E, CDK2, HIF1 alpha & beta-actin was detected by western blot. n = 3. (C–F) Cells were pretreated with trehalose (10 μM), rapamycin (10 nM), PF4708671 (10 μM) & cycloheximide (4 μM) for 30 min prior to A beta treatment for 24 h. Cyclin D1, CDK4, cyclin E, CDK2 & beta-actin were detected by western blot. n = 3–6. (G) Mouse hippocampal neurons were transfected with hif1 alpha specific- & NT siRNAs for 24 h prior to A beta treatment for 24 h. Samples were blotted with Cyclin D1, CDK4, cyclin E, CDK2 & beta-actin specific antibodies. n = 3–6. (H) Mouse hippocampal neurons were pretreated with trehalose (10 μM) for 30 min & incubated with A beta for 24 h. cyclin D1, CDK4, cyclin E, CDK2, HIF1 alpha & beta-actin were analyzed by western blot. n = 3–6. Data are presented as a mean ± SE. *p < 0.05 vs. control, #p < 0.05 vs. A beta treatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28790888), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Hypoxia induces expression of WNT11 through VHL.(A,B) Higher basal levels of WNT11 protein in Vhl-deleted cells (lenti-Cre infected Vhlf/f). EMSCs isolated from Vhlf/f mouse were infected with lentivirus carrying either GFP gene (for control) or Cre recombinase (for knockout). Non-infected cells were also used as a control. Immunoblot analysis of control or Vhl KO EMSCs treated with 0.1 mM DMOG (A), & EMSCs exposed to air (21% O2) or hypoxia (1% O2) for 24 hrs (B). Laminin, alpha-tubulin, & lamin A/C were used as loading controls, WNT11 normalized to alpha-Tubulin was shown. (C,D) Inactivation of the Vhl gene results in increased Wnt11 mRNA. Wnt11 & Vegf mRNA levels in liver (C) or duodenum (D) were measured by qPCR in Liver-VhlcKO or duodenum-VhlcKO & control mice (n = 5 per group). Values normalized to Tbp mRNA are expressed relative to tissues from control mice. For panels (C,D), values are mean ± s.e.m. *p < 0.05, **p < 0.01. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/srep21520), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - QSCs are hypoxic in the niche & express HIF2A, but not HIF1A.(A) Timeline of in vivo pimonidazole labeling in SC-INTACT mice & representative confocal images of uninjured/resting EDL myofibers (n >50 myofibers from n = 3 mice) showing that nmGFP+ QSCs were pimonidazole+. Scale bars: 50 μm & 10 μm (insets). Inset images show that pimonidazole signals were relatively enriched in the cytoplasm of QSCs. Arrowheads indicate a QSC; asterisks indicate a myonucleus. (B) Percentage of pimonidazole+ QSCs. (C) Timeline of in vivo CCI-103F labeling in C57BL/6 mice & representative images of uninjured/resting EDL myofibers (n >50 myofibers from 3 mice) showing that nmGFP+ QSCs were CCI-103F+. Arrowheads indicate a QSC; asterisks indicate a myonucleus. Scale bar: 20 μm. (D) Percentage of CCI-103F+ QSCs. (E & F) Representative images of uninjured/resting EDL myofibers from C57BL/6 mice (n >50 myofibers from 6 mice/group) showing that most QSCs were HIF2A+, but HIF1A–. Scale bars: 10 μm. (G) Percentage of HIF1A+ & HIF2A+ QSCs. Data represent the mean ± SEM. Image collected & cropped by CiteAb from the following publication (https://www.jci.org/articles/view/96208), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - FDFT1 inhibits glycolysis through suppressing AKT-mTOR-HIF1 alpha pathway in CRC.aFDFT1 overexpression reduced glucose uptake in CT26 cells. bFDFT1 overexpression decreased lactate production via glycolysis in CT26 cells. c ECAR reduced when FDFT1 overexpressed in CT26 cells. d OCR increased when FDFT1 overexpressed in CT26 cells. eFDFT1 knockdown increased glucose uptake in CT26 cells. fFDFT1 knockdown increased lactate production via glycolysis in CT26 cells. g ECAR increased when FDFT1 knocked down in CT26 cells. h OCR decreased when FDFT1 knocked down in CT26 cells. i, mFDFT1 overexpression inhibited protein & mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, & LDHA, in CT26 cells. j, mFDFT1 knockdown increased protein & mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, & LDHA, in CT26 cells. k, nFDFT1 overexpression decreased protein & mRNA expression of AKT, mTOR, & HIF1 alpha. l, nFDFT1 knockdown increased protein & mRNA expression of AKT, mTOR, & HIF1 alpha. o Photograph of dissected tumors (the first line: normal diet, second line: FMD + glucose, third line: FMD, n = 5). p The tumor volumes measured every 2 days after 13th day. The FMD + glucose group can reverse tumor growth inhibition induced by FMD (n = 5; ns: P = 0.1838; P = 0.0001). q The protein level of FDFT1 & mTOR in dissected tumor samples from normal diet group, FMD group & FMD + glucose group measured by western blotting. r The glucose level in these three groups. Error bars, mean ± SD, data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared w/ control group (or normal diet group). #P < 0.05, ##P < 0.01. Image collected & cropped by CiteAb from following publication (https://pubmed.ncbi.nlm.nih.gov/32313017), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Role of HIF-1 alpha, FOXO3 in hypoxia-induced BNIP3 expression. (A) UCB-hMSCs were incubated with various durations of hypoxia (0–48 h). The protein expressions of HIF-1 alpha & beta-actin were detected by western blot. n = 4. (B) UCB-hMSCs were pretreated with NAC (5 mM) for 30 min prior to hypoxia incubation for 24 h. The protein expressions of HIF-1 alpha, lamin A/C & beta-tubulin in non-nuclear & nuclear fractionized cell samples were assessed by using western blot. n = 3. (C) UCB-hMSCs were immuno-stained with HIF-1 alpha & PI (magnification ×600). Scale bars, 37.5 µm. (D) HIF1A siRNA or NT siRNA was transfected to cells prior to hypoxia treatment for 24 h. The mRNA expression of BNIP3 was analyzed by qPCR. n = 6. (E) The protein expressions of BNIP3 & HIF-1 alpha were detected by western blot. n = 4. (F) NAC (5 mM) was pretreated to UCB-hMSCs prior to hypoxia treatment for 24 h. FOXO3, lamin A/C & beta-tubulin proteins expressions were assessed by western blot. n = 3. (G) FOXO3 siRNA transfected to UCB-hMSCs prior to hypoxia treatment for 24 h. The FOXO3 mRNA expression was measured by qPCR. n = 6. (H) BNIP3, FOXO3 & beta-actin expressions were detected by western blot. n = 3. Western blot data were normalized by beta-actin, & qPCR data were normalized by ACTB mRNA expression level. Lamin A/C & beta-tubulin were used as nuclear & non-nuclear protein controls, respectively. Quantitative data are presented as a mean ± S.E.M. All blots & confocal images are representative. *p < 0.05 versus control, #p < 0.05 versus hypoxia. Image collected & cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213231717303804), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - ROS overproduction mediated apoptosis induced by combined treatment of DDP & HIF-1 alpha in PC-3 xenografts & cell culture. (a) ROS monitored by DHE (red) & nuclei by DAPI (blue) staining in PCa xenografts (scale bars, 50 µm). (b) MDA formation of PCa xenografts was examined after various treatments (c,d). PC-3 cells were treated with DDP, si-HIF-1 alpha plasmid, or both, in the presence or absence of NAC (5 mM) or DHLA (0.25 mM) for 24 h. Total lysates (c) & culture media (d) were used to detect cellular H2O2 level. (e,f) Western analysis for HIF-1 alpha as well as cleaved caspase-3 & PARP in PC-3 cells following various treatments. Data were presented as mean ± SD of three independent experiments. *p < 0.05 versus control group; #p < 0.05 versus si-HIF-1 alpha or DDP group; $p < 0.05 versus DDP/si-HIF-1 alpha group. The original blots are presented in Supplementary Figure 8. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-07973-4), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - miR-128 downmodulation regulates MAFG increase in hypoxic condition both in vitro & in vivo. C2C12 cells exposed to hypoxia (2% O2) for 4 h, & miR-128 levels (a) determined by RT-qPCR on total RNAs after normalization w/ the small RNU6. Data are expressed as relative to values obtained in untreated control which set equal to 1. Each column in the panels represents the mean ± SD of 3 independent experiments. ∗p < 0.05. Western blotting analysis (b) of HIF-1 alpha, MAFG, & HMOX-1 performed on total extracts from the same cells, as described in Materials & Methods. Tubulin used as a loading control. mRNA levels of HMOX-1 & x-CT genes (c) determined by RT-qPCR on total RNAs, & relative changes calculated by comparing treated cells versus control, after normalization w/ c-ABL. Data reported as relative to values obtained in normoxia, which set equal to 1. Each column in the panel represents the mean ± SD of 3 independent experiments. ∗p < 0.05. Representative laser Doppler analysis (d) of blood flow before & 4 hours after (post 4 h) hindlimb ischemia procedure (n = 3-4 mice/group). Representative Western blot analysis & densitometric analysis (e) of MAFG & HMOX-1 protein levels performed on hindlimb muscle lysates from littermates after sham procedure (sham) or not-ischemic limb (NIL) & ischemic limb (IL) 4 h after femoral artieriectomy. Tubulin protein levels used as loading controls (∗p < 0.05 versus sham; n = 3-4 hindlimb/group). miR-128 levels (f) determined by RT-qPCR on total RNAs after normalization w/ the small RNU6. The values obtained in not-ischemic limb (NIL) set equal to 1. The data are expressed as the mean ± standard error & are representative of 3 independent experiments. ∗p < 0.05. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29138682), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Effect of ENMD-1198 on HIF-1 alpha & VEGF expression in HCC cells. To determine effects of ENMD-1198 on nuclear HIF-1 alpha expression, HUH-7 cells were incubated for 16 h ± ENMD-1198 & subsequently exposed to hypoxia (1% O2, 4 h). A) Western blot analysis of whole protein showed that ENMD-1198 effectively blunted hypoxic induction of HIF-1 alpha protein. B) In addition, treatment with ENMD-1198 down-regulated constitutive VEGF-A mRNA levels in HCC cells (HepG2), as measured by real-time PCR (* P < 0.01) (n = 3/group). VEGF-A mRNA expression is normalized to beta-actin. C) ELISA analysis for VEGF in culture supernatants (HepG2). Hypoxia (20 h, 1% O2) markedly increased VEGF protein. ENMD-1198 treatment lowered VEGF secretion under hypoxic conditions (*P < 0.05). D) Western blot analysis for VEGF. Cells were incubated under either non-hypoxic, or hypoxic conditions ± ENMD-1198. Treatment with ENMD-1198 slightly lowered the hypoxic induction of VEGF, as determined by densitometry. Bars: SEM. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/18651980), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Upregulation of HIF-1 alpha in human PCa. (a) HIF-1 alpha protein was detected by western blot in nonmalignant (RWPE-1 & BPH1) & PCa cell lines (PC-3, DU145, LNCaP, & 22RV1) as indicated. (b–d) Total RNA extracted from RWPE-1, BPH1, PC-3, DU145, LNCaP, & 22RV1 cells was subjected to qRT-PCR for HIF-1 alpha (b), VEGF (c) & GLUT4 (d). (e) The HIF-1 alpha promoter-driven reporter (firefly luciferase) & a control vector (Renilla luciferase) were co-transfected into RWPE-1, BPH1, PC-3, DU145, LNCaP, & 22RV1 cells for measurement of luciferase activity. HIF-1 alpha promoter activity was calculated as a ratio of firefly to Renilla activity. (f) Human normal & malignant tissue (Gleason score 9) sections were probed with HIF-1 alpha antibody (scale bars, 100 µm). (g) Protein expression of HIF-1 alpha, VEGF, & GLUT4 were examined with western blot, in PC-3, DU145, & LNCaP cells after various treatments as indicated. Data are expressed as mean ± SD of seven independent experiments. $p < 0.05 versus RWPE-1 or BPH1 cells or normal tissue. *p < 0.05 versus control group. #p < 0.05 versus si-HIF-1 alpha or DDP group. Original blots are shown in Supplementary Figure 5. C: Ctrl; D: DDP; S: si-HIF-1 alpha; D/S: DDP/si-HIF-1 alpha. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-07973-4), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Cx43-overexpressing cells regulate the expression of HIF-1 alpha, vascular endothelial growth factor (VEGF), & the proliferation of endothelial cells. (a) The expression of Cx43 & HIF-1 alpha was measured in Cx43-overexpressing cells by Western blot analysis. beta-actin expression served as loading controls for & total protein. Inserted values indicated relative proteins expression in comparison with beta-actin; (b) Cx43-overexpressing cells were cotransfected with pCLNCX-6× HRELuc & pTCYLacZ plasmids. At 6 h post-transfection, their luciferase activities were determined & normalized with beta-gal activity. Data shown were the mean ± SD (n = 4); (c) The conditioned medium of Cx43-overexpressing cells was measured by ELISA. Data shown were the mean ± SD (n = 4); & (d) The conditioned medium of Cx43-overexpressing cells reduced the proliferation of endothelial cells. The HEMC-1 cells treated with conditioned medium of Cx43-overexpressing cells were examined the proliferation activity. Cell viability was assessed by the WST-1 assay. Data shown were the mean ± SD (n = 4). * p < 0.05; *** p < 0.001. Image collected & cropped by CiteAb from the following publication (http://www.mdpi.com/1422-0067/16/1/439), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Prolonged hypoxia increases glutamine metabolism in PDAC cells, & HIF‐2 alpha can promote non‐canonical glutamine metabolism in chronic hypoxic conditions. (A) Time course of glutamine consumption at 1%, 3% or 20% O2, each time data point is an average of triplicate experiments. (B) Panc‐1 & Capan‐2 were incubated for 48 hrs at 1%, 3% or 20% O2, GLS1, GOT1 & GOT2 protein were measured by Western blot. beta‐Actin was used as loading control. (C) Panc‐1 & Capan‐2 were incubated for 48 hrs at 1%, 3% or 20% O2, GLS1, GOT1 & GOT2 mRNA were measured by qRT‐PCR. Data are presented as mean ± S.D. from three independent experiments. (D & F) Si‐HIF‐2 alpha‐transfected Panc‐1 & Capan‐2 cultured at 1% or 3%O2 for 48 hrs. The level of HIF‐2 alpha & glutamine metabolism enzymes mRNA & protein were determined by qRT‐PCR (mRNA) & Western blot (protein), & beta‐actin was used as loading control. Data are presented as mean ± S.D. from three independent experiments. *P < 0.05, **P < 0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28544376), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF1 alpha & PFKFB3 are protective against hIAPP mediated beta-cell death. a Representative immunoblotting (left) & quantification (right) of the whole cell extract (WCE) from asynchronous INS 832/13 cells treated with either HIF1 alpha inhibitor KC7F2 (10 μM) or PFKFB3 siRNA (75 nM) for 56 h versus control. Cells were transduced with hIAPP adenovirus (75 MOI) or LacZ adenovirus as control (75 MOI) for last 32 h of culture. DMSO or CTRL siRNA, respectively were added to LacZ or hIAPP to equal DMSO or PFKFB3 siRNA used in experimental groups. GAPDH was used as a loading control. Data are presented as mean ± SEM, n = 4, *p < 0.05, **p < 0.01. b Quantification of DNA content distribution after FACS analysis of INS 832/13 cells treated as described in (a). Data are the mean ± SEM, n = 4, *p < 0.05, **p < 0.01. c The frequency of TUNEL positive INS 832/13 cells treated as described in (a). Data are the mean ± SEM, n = 4 in (a) & (b), & n = 3 independent experiments in (c). Statistical significance was analyzed by one-way ANOVA test with Tukey’s post-test (*p < 0.05, **p < 0.01). See Supplementary Fig. 10 for additional supportive data Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31213603), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Immunohistochemical & Western blot analysis of HIF-1 alpha & VEGF expression in OVA-challenged mice.(A & B) Immunostaining for HIF-1 alpha & VEGF in nasal mucosa following the last challenge. Positive HIF-1 alpha signals are brown & are predominantly nuclear & VEGF is predominantly cytoplasmic (magnification 400×, scale bar = 20 µm). (C & D) Representative Western blot analysis showing HIF-1 alpha & VEGF (with beta-actin as a loading control) expression in the nasal mucosa 24 h after the last challenge. All densitometric analyses are presented as the relative ratio of each molecule to beta-actin & the ratio in negative control mice was set to 100. Values represent means±SEM (n = 6 mice). *Significantly different from negative control, p<0.05; #significantly different from positive control, p<0.05. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0048618), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - DC101, evofosfamide, & HIF-1 alpha shRNA treatment of HT1080 sarcomasA. Western blot of HT1080 cells stably transduced with HIF-1 alpha shRNA (sh.HIF-1 alpha) or scrambled control shRNA (sh.Scr) & grown in 21% oxygen or 1% oxygen. beta-actin serves as loading control. B. Growth of HT1080 cells stably transduced with sh.HIF-1 alpha or sh.Scr in athymic nude mice. Groups were treated with control IgG 20 mg/kg 3 times per week, DC101 20 mg/kg 3 times per week, and/or evofosfamide (Evo) 50 mg/kg 5 times per week. C. Photos of H&E staining, cleaved caspase 3 IF, cleaved caspase 3 & CD31 IF, CD31 IHC, pimonidazole IHC, & HIF-1 alpha IHC. D. Graphs of microvessel density, pimondizale area, & nuclear HIF-1a expression. Bars represent standard deviation. **p < 0.05 compared to all other groups. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.10212), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - WNT11 is induced by hypoxia or hypoxic mimetics in different cell types.(A) Increased Wnt11 mRNA in EMSC adipocytes (Day 12) after hypoxia-mimetic treatments. EMSC adipocytes were treated with CoCl2 (0.1 mM), DFO (0.1 mM) or DMOG (0.1 mM) for 24 hrs. Values were normalized to Tbp mRNA & are expressed relative to control (n = 3). (B,C) Increased Wnt11 mRNA by hypoxia in EMSC preadipocytes & adipocytes (Day 0–12 after differentiation) (B), & C2C12 myoblast & myocyte (Day 0 & 8 after differentiation) (C). Wnt11 mRNA was assessed by quantitative PCR in cells exposed to air (21% O2) or hypoxia (1% O2) for 24 hrs. (n = 4). Values were normalized to Tbp mRNA & are expressed relative to 21% O2 samples (left panel). (D) Immunoblot analyses of HeLa cells under normal air or hypoxia for 24 hrs. (E,F) Induction of Wnt11 by increasing concentrations of DMOG in MDA-MB-231 cells (E) & 4T1 cells (F). (G) EMSCs treated with 0.1 mM DMOG for the indicated times. Wnt11 & Vegf mRNA expression was measured by qPCR & normalized to Tbp mRNA (n = 4). (H) WNT11 protein levels after DMOG treatment normalized to alpha-Tubulin (upper panel; n = 4). Representative immunoblots of EMSCs treated with 0.1 mM DMOG for the indicated times (Lower panel). (I) Protein expression in MDA-MB-231 cells treated with 0.1 mM DMOG. (J) Induction of Wnt11 promoter activity by hypoxia or hypoxia mimetics. pGL3-Wnt11 promoter plasmid was transfected into C2C12 cells. Cells were incubated with DMOG (left panel, n = 4) or under 21% O2 or 1% O2 (right panel, n = 8) for 24 hrs. For panels (A–C,G,H,J), values are mean ± s.e.m. *p < 0.05, **p < 0.01. For panels of immunoblotting, laminin, alpha-tubulin, & ERK were used as loading controls, WNT11 normalized to alpha-Tubulin was shown. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/srep21520), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - CAPE inhibits MCT-induced HIF-1 alpha expression & PDGF-BB production in rats. (A) Rats were treated with PBS or CAPE (5 or 10 mg/kg) from days 14–28 after MCT treatment 14 days (60 mg/kg). Preparation of lung tissues was analyzed by Western blot to determine the levels of HIF-1 alpha protein. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)was used as a loading control. (B) Serum levels of PDGF-BB in rat was assayed by an ELISA kit. (C) Representative images of immunohistochemical staining for HIF-1 alpha in lung sections from the rats. Scale bars, 50 μm. (D) Immunofluorescence staining of alpha-smooth muscle actin ( alpha-SMA) & HIF-1 alpha in lung sections from rats. Scale bars, 50 μm. Data in (A,B) are expressed as mean ± SEM of three independent experiments. *** p < 0.01, as compared with the PBS group. ### p < 0.01, as compared with the rats exposed to MCT alone. Results are representative of three rats per experimental group. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30909527), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - mTOR expression is inversely correlated with FDFT1 expression.aFDFT1 overexpression inhibited the protein level of mTOR, whereas FDFT1 knockdown increased the protein level of mTOR in CT26 & SW620 cells. bFDFT1 expression & mTOR expression were negatively correlated in CRC patient samples. Scale bar: 200 µm. c The mTOR silencing efficiency of the siRNA in CT26 & SW620 cells was validated by western blotting. d The effect of mTOR silencing on FDFT1 expression level in CT26 was evaluated by western blotting. e The protein level of mTOR when FDFT1 knockdown combined with or without mTOR inhibitor in CT26. f The protein level of pS6k, S6k, pS6, & S6 under the effect of fasting & FDFT1 overexpression in CRC cells. g, h CCK8 proliferation assays showed that the silencing of mTOR decreased the proliferation of CT26 & SW620 cells. k, n The silencing of mTOR reduced glucose uptake & lactate production in CT26 & SW620 cells. i, j, l, o The silencing of mTOR decreased the ECAR & increased the OCR in CT26 & SW620 cells. m The silencing of mTOR decreased the expression of AKT, HIF1 alpha, & proteins encoded by relevant glycolytic genes, such as GLUT1, HK2, LDHA, GPI, PGK1, in CT26 & SW620 cells. p–t Based on TCGA data set analysis, mTOR expression was positively correlated with AKT1, HIF1 alpha, GLUT1, HK2, & LDHA expression. Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the control group. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32313017), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Chromatin Immunoprecipitation: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Chromatin Immunoprecipitation: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha-mediated repression of ENT1 expression in Caki-1 cells under hypoxia. Comparison of HIF-1 alpha & HIF-2 alpha protein expression in human matched renal normal-tumor samples (n=11) (A) & 3 RCC cell lines (786-O, 769-P, Caki-1) under normoxia or hypoxia for 6 h (B). (C) Schematic diagram of two hypoxia response elements (HRE) in the promoter of ENT1. (D) CHIP-qPCR analysis of HIF-1 alpha at the promoter of ENT1 in Caki-1 cells, exposed under normoxia or hypoxia for 48 h. Two different HIF-1 alpha antibodies (610958, BD Biosciences; NB100-105, Novus) were used to pull down HIF-1 alpha. The protein expression level of HIF-1 alpha (E) & mRNA expression level of SLC29A1 (F) in Caki-1 under normoxia or hypoxia & HIF-1 alpha knockout-Caki-1 cells. (G) The expression level of ARNT1 (HIF-1 beta) was detected in Caki-1, transfected with ARNT1 specific siRNAs or NC. Student's t test (two-tailed) was used. (H, I) The expression level of ENT1 in Caki-1, transfected with ARNT1 specific siRNAs or NC, under normoxia or hypoxia. Oneway-ANOVA analysis was used. The results are expressed as mean ± SEM (n=3). ***P<0.001. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32206108), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Upregulation of HIF-1 alpha in human PCa. (a) HIF-1 alpha protein was detected by western blot in nonmalignant (RWPE-1 & BPH1) & PCa cell lines (PC-3, DU145, LNCaP, & 22RV1) as indicated. (b–d) Total RNA extracted from RWPE-1, BPH1, PC-3, DU145, LNCaP, & 22RV1 cells was subjected to qRT-PCR for HIF-1 alpha (b), VEGF (c) & GLUT4 (d). (e) The HIF-1 alpha promoter-driven reporter (firefly luciferase) & a control vector (Renilla luciferase) were co-transfected into RWPE-1, BPH1, PC-3, DU145, LNCaP, & 22RV1 cells for measurement of luciferase activity. HIF-1 alpha promoter activity was calculated as a ratio of firefly to Renilla activity. (f) Human normal & malignant tissue (Gleason score 9) sections were probed with HIF-1 alpha antibody (scale bars, 100 µm). (g) Protein expression of HIF-1 alpha, VEGF, & GLUT4 were examined with western blot, in PC-3, DU145, & LNCaP cells after various treatments as indicated. Data are expressed as mean ± SD of seven independent experiments. $p < 0.05 versus RWPE-1 or BPH1 cells or normal tissue. *p < 0.05 versus control group. #p < 0.05 versus si-HIF-1 alpha or DDP group. Original blots are shown in Supplementary Figure 5. C: Ctrl; D: DDP; S: si-HIF-1 alpha; D/S: DDP/si-HIF-1 alpha. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-07973-4), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Upregulation of HIF-1 alpha in the spleen restricts CD8+ T cell expansion.(A-B) Mice were infected with 2x107 amastigotes intravenously & euthanized at various time points after infection. HIF-1 alpha expression in splenocytes was assessed by real-time PCR (A) & immunoblot analysis (B). (C-F) 2x104 OT-I CD8+ T cells were adoptively transferred into recipient mice a day prior to infection with ovalbumin-transgenic (PINK) L. donovani amastigotes. (C) Graphs represent the average number of OT-I CD8+ T cells found in the spleen from Hif1aflox/flox & Hif1a+/-mice at d7 & d14 p.i.. (D) Percentage of OT-I CD8+ T cells at d7 & d14 p.i.. (E) Percentage of gated OT-I CD8+ expressing CD62Llo/int & negative for CD127. (F) Percentage of OT-I CD8+ T cells that were CD44+ CD127+CD62Lhi. All data represent mean ± SEM of one of 3 independent experiments, n = 5. * denotes p<0.05 & ** denotes p<0.01. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.ppat.1004938), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Ccl7 is a HIF1 alpha direct target gene.a qPCR analysis of Ccl7 mRNA expression in Hif1afl/fl & Hif1a deltaSMC VSMCs treated with vehicle, CoCl2, normoxia, or hypoxia (2% O2) for 6, 12, & 24 h. *P < 0.05, **P < 0.01, ***P < 0.001, n = 6 (independent experiments) per group. VSMCs isolated from Hif1afl/fl mice & Hif1a deltaSMC mice were infected with oxygen-stable HIF1 alpha-expressing lentivirus, & then treated with Ang II for 24 h, bCcl7 mRNA was measured by qPCR & c CCL7 protein was detected by ELISA. d qPCR analysis of Ccl7 mRNA expression in vehicle or Ang II-treated Hif2afl/fl & Hif2a deltaSMC VSMCs. e Schematic diagram of the mouse Ccl7 promoter illustrating the HREs in the regulatory region; the upstream regions were numbered in relation to the transcription initiation site. f Luciferase-reporter constructs under the control of the mouse Ccl7 promoter. HEK293T human embryonic kidney cells transiently transfected with the luciferase construct, & cotransfected with empty vector or HIF1a expression plasmids. Standard dual-luciferase assays were performed. EV, empty vector. **P < 0.01, n = 3. g, h ChIP assays of vehicle or Ang II-treated wild-type VSMCs using HIF1 alpha or HIF2 alpha antibodies. Data were normalized to input. *P < 0.05, **P < 0.01, n = 6 per group. i, j ChIP assays of vehicle or Ang II-treated Hif1afl/fl & Hif1a deltaSMC VSMCs using HIF1 alpha antibody. Data were normalized to input. *P < 0.05, **P < 0.01, n = 6 per group. Statistical significance was determined by one-way ANOVA test followed by the unpaired t-test Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31320613), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - WNT11 is induced by hypoxia or hypoxic mimetics in different cell types.(A) Increased Wnt11 mRNA in EMSC adipocytes (Day 12) after hypoxia-mimetic treatments. EMSC adipocytes were treated with CoCl2 (0.1 mM), DFO (0.1 mM) or DMOG (0.1 mM) for 24 hrs. Values were normalized to Tbp mRNA & are expressed relative to control (n = 3). (B,C) Increased Wnt11 mRNA by hypoxia in EMSC preadipocytes & adipocytes (Day 0–12 after differentiation) (B), & C2C12 myoblast & myocyte (Day 0 & 8 after differentiation) (C). Wnt11 mRNA was assessed by quantitative PCR in cells exposed to air (21% O2) or hypoxia (1% O2) for 24 hrs. (n = 4). Values were normalized to Tbp mRNA & are expressed relative to 21% O2 samples (left panel). (D) Immunoblot analyses of HeLa cells under normal air or hypoxia for 24 hrs. (E,F) Induction of Wnt11 by increasing concentrations of DMOG in MDA-MB-231 cells (E) & 4T1 cells (F). (G) EMSCs treated with 0.1 mM DMOG for the indicated times. Wnt11 & Vegf mRNA expression was measured by qPCR & normalized to Tbp mRNA (n = 4). (H) WNT11 protein levels after DMOG treatment normalized to alpha-Tubulin (upper panel; n = 4). Representative immunoblots of EMSCs treated with 0.1 mM DMOG for the indicated times (Lower panel). (I) Protein expression in MDA-MB-231 cells treated with 0.1 mM DMOG. (J) Induction of Wnt11 promoter activity by hypoxia or hypoxia mimetics. pGL3-Wnt11 promoter plasmid was transfected into C2C12 cells. Cells were incubated with DMOG (left panel, n = 4) or under 21% O2 or 1% O2 (right panel, n = 8) for 24 hrs. For panels (A–C,G,H,J), values are mean ± s.e.m. *p < 0.05, **p < 0.01. For panels of immunoblotting, laminin, alpha-tubulin, & ERK were used as loading controls, WNT11 normalized to alpha-Tubulin was shown. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/srep21520), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - CAPE inhibits MCT-induced HIF-1 alpha expression & PDGF-BB production in rats. (A) Rats were treated with PBS or CAPE (5 or 10 mg/kg) from days 14–28 after MCT treatment 14 days (60 mg/kg). Preparation of lung tissues was analyzed by Western blot to determine the levels of HIF-1 alpha protein. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)was used as a loading control. (B) Serum levels of PDGF-BB in rat was assayed by an ELISA kit. (C) Representative images of immunohistochemical staining for HIF-1 alpha in lung sections from the rats. Scale bars, 50 μm. (D) Immunofluorescence staining of alpha-smooth muscle actin ( alpha-SMA) & HIF-1 alpha in lung sections from rats. Scale bars, 50 μm. Data in (A,B) are expressed as mean ± SEM of three independent experiments. *** p < 0.01, as compared with the PBS group. ### p < 0.01, as compared with the rats exposed to MCT alone. Results are representative of three rats per experimental group. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30909527), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Role of HIF-1 alpha, FOXO3 in hypoxia-induced BNIP3 expression. (A) UCB-hMSCs were incubated with various durations of hypoxia (0–48 h). The protein expressions of HIF-1 alpha & beta-actin were detected by western blot. n = 4. (B) UCB-hMSCs were pretreated with NAC (5 mM) for 30 min prior to hypoxia incubation for 24 h. The protein expressions of HIF-1 alpha, lamin A/C & beta-tubulin in non-nuclear & nuclear fractionized cell samples were assessed by using western blot. n = 3. (C) UCB-hMSCs were immuno-stained with HIF-1 alpha & PI (magnification ×600). Scale bars, 37.5 µm. (D) HIF1A siRNA or NT siRNA was transfected to cells prior to hypoxia treatment for 24 h. The mRNA expression of BNIP3 was analyzed by qPCR. n = 6. (E) The protein expressions of BNIP3 & HIF-1 alpha were detected by western blot. n = 4. (F) NAC (5 mM) was pretreated to UCB-hMSCs prior to hypoxia treatment for 24 h. FOXO3, lamin A/C & beta-tubulin proteins expressions were assessed by western blot. n = 3. (G) FOXO3 siRNA transfected to UCB-hMSCs prior to hypoxia treatment for 24 h. The FOXO3 mRNA expression was measured by qPCR. n = 6. (H) BNIP3, FOXO3 & beta-actin expressions were detected by western blot. n = 3. Western blot data were normalized by beta-actin, & qPCR data were normalized by ACTB mRNA expression level. Lamin A/C & beta-tubulin were used as nuclear & non-nuclear protein controls, respectively. Quantitative data are presented as a mean ± S.E.M. All blots & confocal images are representative. *p < 0.05 versus control, #p < 0.05 versus hypoxia. Image collected & cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213231717303804), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Cx43-knockdown cells regulate the expression of HIF-1 alpha, c-Src, VEGF & the proliferation of endothelial cells. (a) The expression of Cx43, c-Src, & HIF-1 alpha was measured in Cx43-knocknown cells by Western blot analysis. beta-actin expression served as loading controls for & total protein. Inserted values indicated relative protein expression in comparison with beta-actin; (b) Cx43-knockdown cells were cotransfected with pCLNCX-6× HRELuc & pTCYLacZ plasmids. Then, cells were treated with 100 ng/μL PP2 or DMSO for 1 h. At 6 h post-transfection, their luciferase activities were determined & normalized with beta-gal activity. Data shown were the mean ± SD (n = 4); (c) The conditioned medium of Cx43-knockdown cells was measured by ELISA. Data shown were the mean ± SD (n = 4); (d) The conditioned medium of Cx43-knockdown cells reduced the proliferation of endothelial cells. The HEMC-1 cells treated with conditioned medium of Cx43-knockdown cells were examined for proliferation activity. Cell viability was assessed by the WST-1 assay. Data shown were the mean ± SD (n = 4). * p < 0.05; ** p < 0.01; *** p < 0.001. Image collected & cropped by CiteAb from the following publication (http://www.mdpi.com/1422-0067/16/1/439), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Involvement of CBP in BNIP3 expression regulated by HIF-1 alpha & FOXO3 under hypoxia. (A) UCB-hMSCs were incubated with hypoxia condition for 24 h. Co-immunoprecipitation of HIF-1 alpha & FOXO3 with IgG & CBP were shown in left panel. IgG was used as a negative control. The total protein expressions of HIF-1 alpha, FOXO3, CBP & beta-actin in lysate were shown in right panel. n = 3. (B) CBP (20 μM) was pretreated to UCB-hMSCs, & cells were incubated with hypoxia for 24 h. The BNIP3 mRNA expression level was analyzed by qPCR. n = 6. (C) BNIP3 & beta-actin protein expressions were analyzed by western blot. Data represent mean ± S.E. n = 4. (D, E) Sample DNA was immuno-precipitated with RNA polymerase, IgG, HIF-1 alpha & FOXO3 specific antibodies. CHIP (top panel) & lysate (bottom panel) samples were amplified with the primers of GAPDH & BNIP3 promoters. Quantitative CHIP data was analyzed by qPCR, & shown in the right panel. n = 4. Western blot data were normalized by beta-actin, & qPCR data were normalized by ACTB mRNA expression level. Quantitative data are presented as a mean ± S.E.M. All blot images are representative. *p < 0.05 versus control, #p < 0.05 versus hypoxia. Image collected & cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213231717303804), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Precision quantitative image analysis workflow for the characterization of functional ITH.(a) Representative areas with positive immunohistochemical (IHC) staining for HIF-1 alpha, HIF-2 alpha, phospho-mTOR S2448, phospho-S6RP S235/236 & Ki-67. Only nuclear HIF-1 alpha & HIF-2 alpha expression was included in the analysis. Scale bar, 100 μm. (b) Overview of the quantitative IHC analysis process using a representative image of a whole-slide scanned pT1M1 tumour after phospho-mTOR S2448 staining. The image was overlaid with a virtual grid consisting of 1 mm2 squares that defines the tumour regions to be analysed. Non-cancerous tissue was subtracted manually (dark grey) & a positive pixel count was performed on the corrected area. Positivity per square was expressed as percentage of positive pixels. If a square contained non-tumorous tissue, the positivity was normalized to the corrected square area. Computed values were depicted as a heat map. Coloured squares represent tumour, whereas open squares indicate completely non-tumorous areas. Squares defined as peripheral tumour zone are marked by a black line. Scale bar, 1 cm. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27291893), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - ROS overproduction mediated apoptosis induced by combined treatment of DDP & HIF-1 alpha in PC-3 xenografts & cell culture. (a) ROS monitored by DHE (red) & nuclei by DAPI (blue) staining in PCa xenografts (scale bars, 50 µm). (b) MDA formation of PCa xenografts was examined after various treatments (c,d). PC-3 cells were treated with DDP, si-HIF-1 alpha plasmid, or both, in the presence or absence of NAC (5 mM) or DHLA (0.25 mM) for 24 h. Total lysates (c) & culture media (d) were used to detect cellular H2O2 level. (e,f) Western analysis for HIF-1 alpha as well as cleaved caspase-3 & PARP in PC-3 cells following various treatments. Data were presented as mean ± SD of three independent experiments. *p < 0.05 versus control group; #p < 0.05 versus si-HIF-1 alpha or DDP group; $p < 0.05 versus DDP/si-HIF-1 alpha group. The original blots are presented in Supplementary Figure 8. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-07973-4), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Regulation of OA development by HIF-1 alpha. Plain radiographs of the entire bodies at 8 weeks of age (A), & knee joints 8 weeks after OA surgery (B) of Cntl & cKO littermates. Scale bars, 10 mm & 1 mm, respectively. (C) Safranin O & TUNEL staining’s of Cntl & KO knee joints at 8 weeks after OA surgery (n = 9 & 11, respectively). Tamoxifen induction was performed at 7 weeks. Inset boxes in top images indicate the regions of high magnification Safranin O & TUNEL images. Inset boxes in middle images indicate the regions of immunofluorescence in (G). White dotted lines in TUNEL images indicate the cartilage area. Scale bars, 400 µm & 100 µm, respectively. (D) OARSI (Osteoarthritis Research Society International) score of OA development. Lines show the mean ± SD. *P < 0.05. (E) Rate of TUNEL-positive cells in the cartilage area. Bars show the mean ± SD. *P < 0.05. (F) mRNA levels of Hif1a in articular cartilage tissues of Cntl & KO. Bars show the mean ± SD. *P < 0.05. (G) Immunofluorescence of Hif1a, Mmp13, Hif2a, I kappaB alpha, Ser 32 & 36 dual phosphorylated I kappaB alpha, Rela, & C1qtnf3. Scale bars, 20 µm. The percentage of positive cells in the immunofluorescence is shown below. *P < 0.05. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32214220), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - A beta facilitates HIF1 alpha synthesis & autophagy inhibition via mTOR activation. (A) SK-N-MC cells exposed to A beta (5 μM) for 0–48 h. HIF1 alpha & beta-actin expression analyzed by WB. n = 3. (B) Cells pretreated w/ NAC (1 mM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expression analyzed by WB. n = 3. (C,E) Cells incubated w/ rapamycin (10 nM) for 30 min prior to A beta treatment for 24 h. Phosphorylation of 4EBP1 (Thr 37/46) & 4EBP1, phosphorylation of p70S6K1 (Thr 389), HIF1 alpha & beta-actin analyzed by WB. n = 6. (D) Protein samples immunoprecipitated by eukaryotic translation initiation factor 4E (eIF4E) antibody-conjugated protein A/G agarose beads. Samples blotted w/ 4EBP1 & eIF4E-specific antibodies. n = 3. (F) Cells exposed to PF4708671 (10 μM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expression detected by WB. n = 6. (G) Cells exposed to cycloheximide (4 μM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expressions detected by WB. n = 6. (H) Cells pretreated w/ rapamycin (10 nM) for 30 min, incubated w/ A beta for 24 h & analyzed by WB w/ LC3, p62 & beta-actin specific antibodies. n = 3–6. (I) LC3 puncta visualized by confocal microscopy. Presented results merged images. Green & red fluorescents indicate LC3 & PI respectively. Scale bars, 50 μm (magnification × 600). (J) Cells pretreated w/ trehalose (10 μM) for 30 min prior to A beta treatment for 24 h. Cytotoxicity measured by MTT assay at an absorbance of 545 nm using a microplate reader. Data present the mean ± SE. n = 6. (K) Cell viability measured by trypan blue exclusion assay. Data presented as a mean ± SE. n = 6. Each blot image presented as representative image. *p < 0.05 vs. control, #p < 0.05 vs. A beta treatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28790888), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - TSA reduces HIF-1 alpha & VEGF expression & up-regulates the expression of PEDF.Real-time PCR (A) & Western blot assays (B) were performed on RPE cells treated with 0–0.5 μM TSA & 150 μM CoCl2. (A) Cells were treated with 0–0.5 μM TSA for 14 h & then co-treated with 150 μM CoCl2 for 6 h for the analysis of gene expression by real-time PCR. Changes in HIF-1 alpha mRNA levels were not statistically significant. CoCl2 causes a fourfold enhancement of VEGF mRNA expression; but at 0.5 μM TSA, the mRNA level of VEGF reduces to less than half of that in cells treated with CoCl2 only. TSA induces a statistically significant increase in the mRNA level of PEDF. (B) Cells were treated with 0–0.5 μM TSA for 18 h & then co-treated with 150 μM CoCl2 for 6 h for Western blot analysis. TSA reduces the CoCl2-induced HIF-1 alpha & VEGF protein levels by 4.3-fold & 5.7-fold, respectively, & up-regulates PEDF protein level by threefold. (C) Densitometry data for Western blot of HIF-1 alpha, VEGF & PEDF. (*: t test p<0.05; **: t test p<0.01). Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0120587), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - A beta facilitates HIF1 alpha synthesis & autophagy inhibition via mTOR activation. (A) SK-N-MC cells exposed to A beta (5 μM) for 0–48 h. HIF1 alpha & beta-actin expression analyzed by WB. n = 3. (B) Cells pretreated w/ NAC (1 mM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expression analyzed by WB. n = 3. (C,E) Cells incubated w/ rapamycin (10 nM) for 30 min prior to A beta treatment for 24 h. Phosphorylation of 4EBP1 (Thr 37/46) & 4EBP1, phosphorylation of p70S6K1 (Thr 389), HIF1 alpha & beta-actin analyzed by WB. n = 6. (D) Protein samples immunoprecipitated by eukaryotic translation initiation factor 4E (eIF4E) antibody-conjugated protein A/G agarose beads. Samples blotted w/ 4EBP1 & eIF4E-specific antibodies. n = 3. (F) Cells exposed to PF4708671 (10 μM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expression detected by WB. n = 6. (G) Cells exposed to cycloheximide (4 μM) for 30 min prior to A beta treatment for 24 h. HIF1 alpha & beta-actin expressions detected by WB. n = 6. (H) Cells pretreated w/ rapamycin (10 nM) for 30 min, incubated w/ A beta for 24 h & analyzed by WB w/ LC3, p62 & beta-actin specific antibodies. n = 3–6. (I) LC3 puncta visualized by confocal microscopy. Presented results merged images. Green & red fluorescents indicate LC3 & PI respectively. Scale bars, 50 μm (magnification × 600). (J) Cells pretreated w/ trehalose (10 μM) for 30 min prior to A beta treatment for 24 h. Cytotoxicity measured by MTT assay at an absorbance of 545 nm using a microplate reader. Data present the mean ± SE. n = 6. (K) Cell viability measured by trypan blue exclusion assay. Data presented as a mean ± SE. n = 6. Each blot image presented as representative image. *p < 0.05 vs. control, #p < 0.05 vs. A beta treatment. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28790888), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Role of HIF-1 alpha, FOXO3 in hypoxia-induced BNIP3 expression. (A) UCB-hMSCs were incubated with various durations of hypoxia (0–48 h). The protein expressions of HIF-1 alpha & beta-actin were detected by western blot. n = 4. (B) UCB-hMSCs were pretreated with NAC (5 mM) for 30 min prior to hypoxia incubation for 24 h. The protein expressions of HIF-1 alpha, lamin A/C & beta-tubulin in non-nuclear & nuclear fractionized cell samples were assessed by using western blot. n = 3. (C) UCB-hMSCs were immuno-stained with HIF-1 alpha & PI (magnification ×600). Scale bars, 37.5 µm. (D) HIF1A siRNA or NT siRNA was transfected to cells prior to hypoxia treatment for 24 h. The mRNA expression of BNIP3 was analyzed by qPCR. n = 6. (E) The protein expressions of BNIP3 & HIF-1 alpha were detected by western blot. n = 4. (F) NAC (5 mM) was pretreated to UCB-hMSCs prior to hypoxia treatment for 24 h. FOXO3, lamin A/C & beta-tubulin proteins expressions were assessed by western blot. n = 3. (G) FOXO3 siRNA transfected to UCB-hMSCs prior to hypoxia treatment for 24 h. The FOXO3 mRNA expression was measured by qPCR. n = 6. (H) BNIP3, FOXO3 & beta-actin expressions were detected by western blot. n = 3. Western blot data were normalized by beta-actin, & qPCR data were normalized by ACTB mRNA expression level. Lamin A/C & beta-tubulin were used as nuclear & non-nuclear protein controls, respectively. Quantitative data are presented as a mean ± S.E.M. All blots & confocal images are representative. *p < 0.05 versus control, #p < 0.05 versus hypoxia. Image collected & cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213231717303804), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF1 alpha protein is suppressed by ED71 but not by 1,25(OH)2D3.(A) Western analysis of Raw264.7 cells cultured in hypoxic conditions with or without 10−7 M of ED71 or 1,25(OH)2D3 (1,25D). (B) Hif1 alpha mRNA levels in Raw264.7 cells cultured in hypoxic conditions were analyzed by realtime PCR in the presence or absence of 10−7 M ED71 or 1,25(OH)2D3. Data represent mean Hif1 alpha expression relative to that of Actb ± SD (n = 5). (C) Levels of VDR transcripts in Raw264.7 cells transfected with shRNA targeting the VDR (shVDR) or control shRNA (Control) were determined by realtime PCR. Data represent mean VDR expression relative to that of Actb ± SD (n = 5). (D) Western analysis of control (shControl) or VDR-suppressed (shVDR#1 or shVDR#2) Raw264.7 transformants cultured in hypoxic conditions with ED71 or 1,25(OH)2D3 (1,25D), both at 10−7 M. (E) M-CSF-dependent Ctsk Cre/Hifflox/flox cells were cultured in normoxic conditions to suppress HIF1 alpha in the presence of M-CSF (50 ng/ml) plus RANKL (25 ng/ml) with either ED71 or 1,25(OH)2D3 (1,25D) both at 10−7M for 4 days. Expression of Ctsk & NFATc1 was then assessed by realtime PCR. Data represent mean Ctsk or NFATc1 expression relative to that of Actb ± SD (n = 5). *P<0.05; **P<0.01; ***P<0.001. Image collected & cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0111845), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - Lack of HIF-P4H-1 increases the amount of p53. (a) Western blot analysis of p53 in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h. (b,c) Western blot analysis of p53 in Hif-p4h-1 & scrambled (Scrm) siRNA transfected HEK293 cells (b) & wt MEFs treated with 50 µM FG4497 for 24 h (c). (d) qPCR analysis of p53 mRNA in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h. (e) Analysis of p53 protein turnover rate. Hif-p4h-1−/− & wt MEFs were treated with 200 μg/ml of cycloheximide for the indicated time points & whole cell lysates were blotted for p53. (f) Western blot analysis of MDM2 in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h. (g) Western blot analysis of p53 & MDM2 in wt MEFs treated with 10 μM nutlin-3a for 24 h with or without overexpression of human HIF-P4H-1 (OE). (h,i) Western blot analysis of HIF1 alpha in wt & Hif-p4h-1−/− MEFs cultured in 21% or 1% O2 for 24 h (h) & in scrambled & Hif-p4h-1 siRNA transfected HEK293 cells (i). (j) Western blot analysis of ubiquitination of p53 in Hif-p4h-1 & scrambled siRNA transfected HEK293 cells. The cells were transfected with HA-ubiquitin & endogenous p53 was immunoprecipitated followed by Western blotting with anti-HA & anti-P53 antibodies. (k) Western blot analysis of p53 in Hif-p4h-1−/− & wt MEFs were treated with or without 10 μM MG132 for 5 h. Data are presented as representative Western blots & as mean ± s.d., n = at least 3 individual MEF isolates or experiments. *P < 0.05, **P < 0.01 & ***P < 0.001, two-tailed Student’s t-test. Unprocessed original scans of blots are shown in Supplementary Fig. 5. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-17376-0), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - NDRG1 was regulated by HIF‐1/2 alpha. (A‐E) The protein expression of the shown genes was detected by Western blot, respectively; beta‐actin was used as the loading control. (A) RCC4 & 786‐O cells (B) were infected with VHL vector (VHL) or empty vector (EV). (C) Caki‐1 cells were transfected with VHL shRNAs (shVHL) or non‐specific control (NC). (D) RCC4 cells were infected with HIF‐1 alpha siRNAs (siHIF‐1 alpha) or non‐specific control (NC). (E) 786‐O cells were transfected with HIF‐2 alpha shRNAs (shHIF‐2 alpha) or non‐specific control (NC). (F) Representative images of HIF‐1 alpha, HIF‐2 alpha & NDRG1 protein expression from the IHC staining of human clear cell renal cell cancer specimens. Patient I: The protein levels of HIF‐1 alpha, HIF‐2 alpha & NDRG1 were low. Patient II: The protein levels of HIF‐1 alpha, HIF‐2 alpha & NDRG1 were medium. Patient III: The protein levels of HIF‐1 alpha, HIF‐2 alpha & NDRG1 were high (scale bar, 50 μm). (G & H) The protein level of NDRG1 is significantly correlated with HIF‐1 alpha (G) & HIF‐2 alpha (H). The protein levels were evaluated according to IHC scores, representing very low (score 0‐2), low (score 3‐4), high (score 5‐8) & strong (score 9‐12). The subjects were divided into four groups according to the IHC scores of HIFs in the tumours. (I) The protein level of NDRG1 is significantly correlated with the average of HIF‐1 alpha & HIF‐2 alpha in ccRCC tissues. The protein levels are evaluated according to IHC scores, representing very low (score < 3), low (score < 5), high (score < 8) & strong (score 8‐12). The subjects were divided into four groups according to the IHC scores of the average of HIF‐1 alpha & HIF‐2 alpha. Significance p‐values & r values according to Spearman's rank correlation Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32537867), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] -
Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - WNT11 is induced by hypoxia or hypoxic mimetics in different cell types.(A) Increased Wnt11 mRNA in EMSC adipocytes (Day 12) after hypoxia-mimetic treatments. EMSC adipocytes were treated with CoCl2 (0.1 mM), DFO (0.1 mM) or DMOG (0.1 mM) for 24 hrs. Values were normalized to Tbp mRNA & are expressed relative to control (n = 3). (B,C) Increased Wnt11 mRNA by hypoxia in EMSC preadipocytes & adipocytes (Day 0–12 after differentiation) (B), & C2C12 myoblast & myocyte (Day 0 & 8 after differentiation) (C). Wnt11 mRNA was assessed by quantitative PCR in cells exposed to air (21% O2) or hypoxia (1% O2) for 24 hrs. (n = 4). Values were normalized to Tbp mRNA & are expressed relative to 21% O2 samples (left panel). (D) Immunoblot analyses of HeLa cells under normal air or hypoxia for 24 hrs. (E,F) Induction of Wnt11 by increasing concentrations of DMOG in MDA-MB-231 cells (E) & 4T1 cells (F). (G) EMSCs treated with 0.1 mM DMOG for the indicated times. Wnt11 & Vegf mRNA expression was measured by qPCR & normalized to Tbp mRNA (n = 4). (H) WNT11 protein levels after DMOG treatment normalized to alpha-Tubulin (upper panel; n = 4). Representative immunoblots of EMSCs treated with 0.1 mM DMOG for the indicated times (Lower panel). (I) Protein expression in MDA-MB-231 cells treated with 0.1 mM DMOG. (J) Induction of Wnt11 promoter activity by hypoxia or hypoxia mimetics. pGL3-Wnt11 promoter plasmid was transfected into C2C12 cells. Cells were incubated with DMOG (left panel, n = 4) or under 21% O2 or 1% O2 (right panel, n = 8) for 24 hrs. For panels (A–C,G,H,J), values are mean ± s.e.m. *p < 0.05, **p < 0.01. For panels of immunoblotting, laminin, alpha-tubulin, & ERK were used as loading controls, WNT11 normalized to alpha-Tubulin was shown. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/srep21520), licensed under a CC-BY license. Not internally tested by Novus Biologicals.


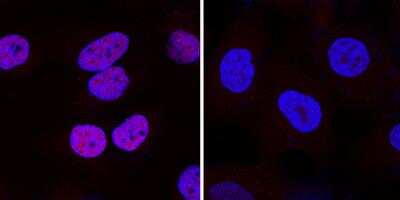
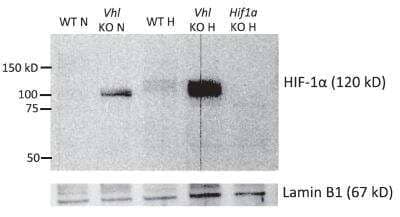
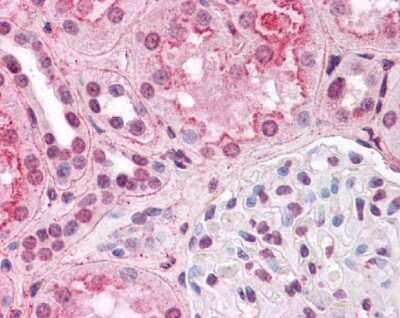
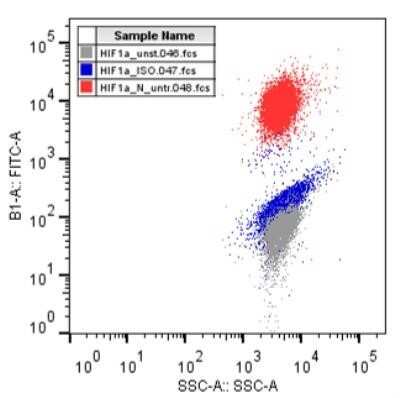
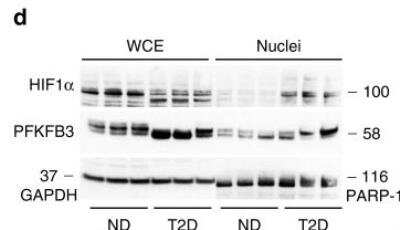
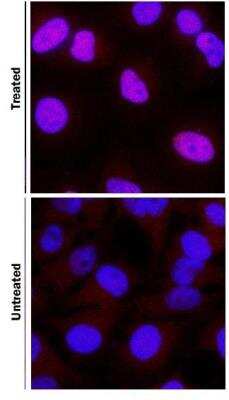
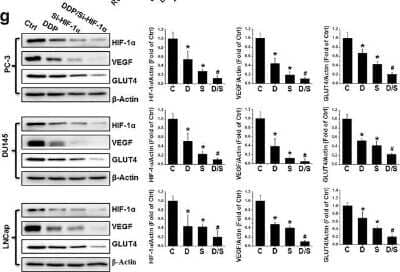
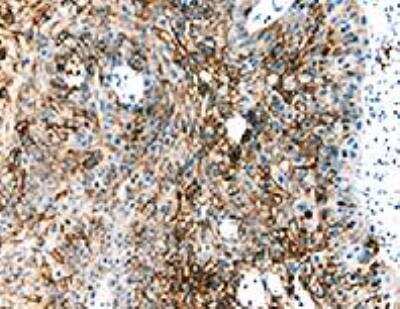
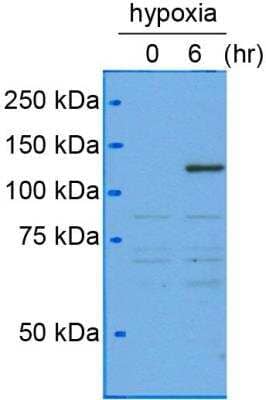
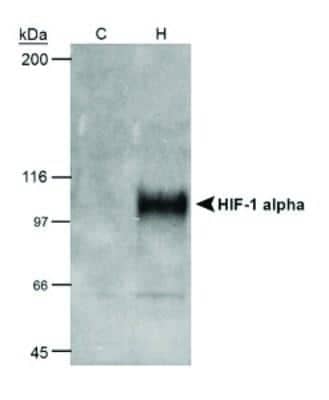
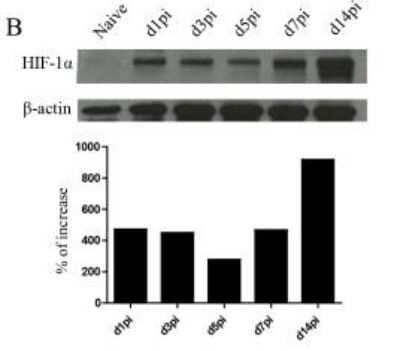
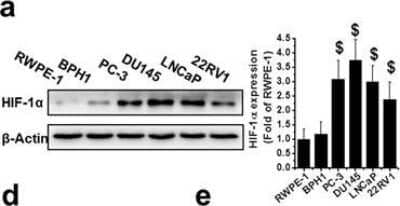
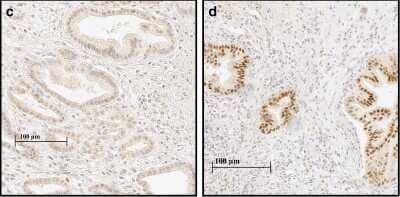
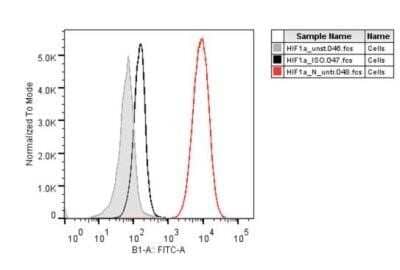
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-271220231261949.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-271220231261920.jpg)
![Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-271220231253311.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-271220231261914.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-271220231285614.jpg)
![Simple Western: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-30820241044555.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-21020242345240.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415284537.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415371973.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415371945.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415371960.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241539593.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415371957.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415395927.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415345343.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415334940.jpg)
![Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241535668.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241535620.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241534367.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415395944.jpg)
![Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415395930.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241535697.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241538246.jpg)
![Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415382424.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415284524.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241529190.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415395911.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415291982.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241517526.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415392560.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415304218.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415175262.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415345342.jpg)
![Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415175253.jpg)
![Immunohistochemistry-Paraffin: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415363727.jpg)
![Chromatin Immunoprecipitation: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535186.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241554198.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415541955.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241553389.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532011.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532019.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532069.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535145.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241612666.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241554534.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535166.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241612678.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024155525.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415525228.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241612625.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532090.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241552521.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415525236.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532060.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241612610.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241554522.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415523943.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415523937.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241612694.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415523933.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535163.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415541970.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532043.jpg)
![Chromatin Immunoprecipitation: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535192.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415525216.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532057.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415541951.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415525295.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024165710.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535122.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415535175.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024155459.jpg)
![Immunohistochemistry: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024168213.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024165739.jpg)
![Immunocytochemistry/ Immunofluorescence: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024155454.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241683940.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241555218.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415541924.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-3102024155458.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241682192.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415541934.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-31020241682138.jpg)
![Western Blot: HIF-1 alpha Antibody (H1alpha67) [NB100-105] - HIF-1 alpha Antibody (H1alpha67)](https://resources.bio-techne.com/images/products/nb100-105_mouse-monoclonal-hif-1-alpha-antibody-h1alpha67-310202415532037.jpg)