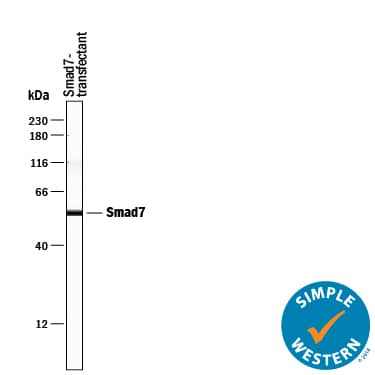
Detection of Human Smad7 by Simple WesternTM.
Simple Western lane view shows lysates ofSf21S. frugiperdainsect ovarian cell line transfected with human Smad7, loaded at 0.2 mg/mL. A specific band was detected for Smad7 at approximately 56 kDa (as indicated) using 10 µg/mL of Mouse Anti-Human/Mouse/Rat Smad7 Monoclonal Antibody (Catalog # MAB2029). This experiment was conducted under reducing conditions and using the 12-230 kDa separation system.
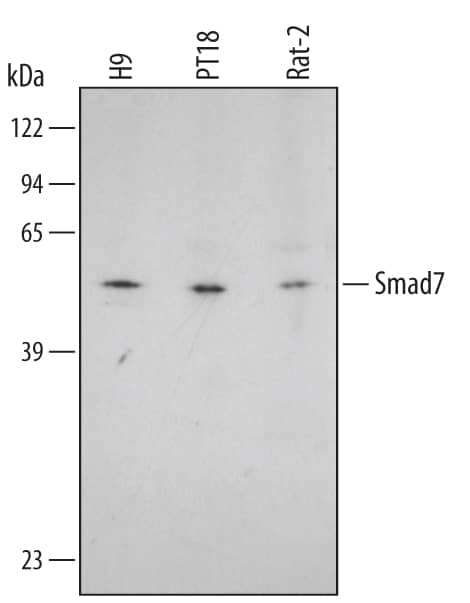
Detection of Human Smad7 by Western Blot
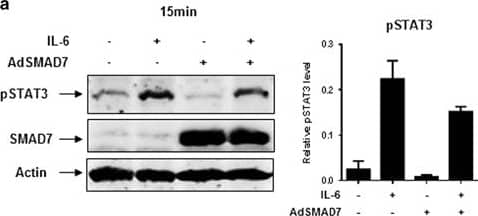
Adenovirus-mediated SMAD7 expression inhibits STAT3 phosphorylation in HuH-7. (a) Ectopic SMAD7 expression inhibits both endogenous and IL-6-induced STAT3 phosphorylation in HuH-7 cells. (b) Ectopic SMAD7 expression inhibits IL-6-induced MCL-1 expression in HuH-7 cells. Quantification of immunoblot analysis was performed of two independent experiments. One result is shown representatively. (c) IL-6 expression levels were investigated by RT–PCR upon ectopic SMAD7 expression. (d) Theoretical pathway connections between SMAD7 and STAT3 in HCC analyzed by cBioPortal analysis (https://www.cbioportal.org/). Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/oncsis201685), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human Smad7 by Western Blot
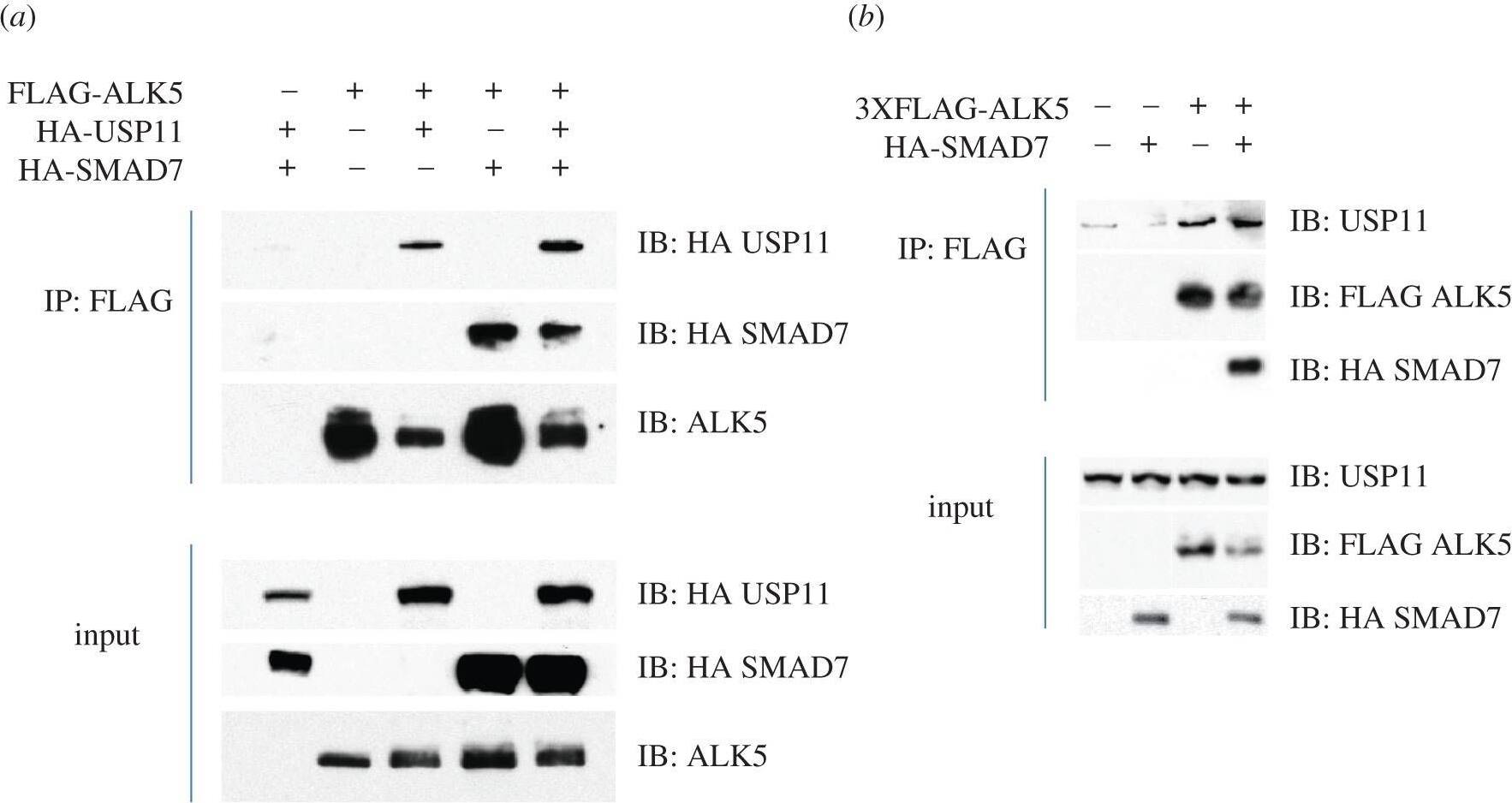
USP11 interacts with ALK5. (a) HEK293 cells were transiently transfected with FLAG-ALK5, HA-USP11 and/or HA-SMAD7, as indicated. Extracts or FLAG IPs were resolved by SDS–PAGE and immunoblotted with antibodies against HA-USP11, HA-SMAD7 and ALK5. (b) HEK293 cells were transiently transfected with 3XFLAG-ALK5, and HA-SMAD7, as indicated. Extracts or FLAG IPs were resolved by SDS–PAGE and immunoblotted with antibodies against endogenous USP11, HA-SMAD7 and 3XFLAG-ALK5. All immunoblots are representative of at least three biological replicates. (c) Lysates from HEK293 cells treated with vehicle or TGF beta (50 pM 45 min) were immunoprecipitated using pre-immune IgG or an ALK5 antibody covalently bound to Dynabeads. IPs, flow-through extracts and lysate inputs were immunoblotted with endogenous USP11, ALK5 and phospho-SMAD2 antibodies. The arrowhead denotes the native molecular weight ALK5. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/22773947), licensed under a CC-BY license. Not internally tested by R&D Systems.
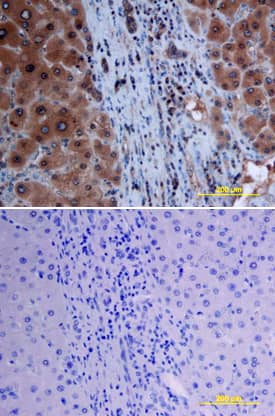
Detection of Mouse Smad7 by Immunohistochemistry
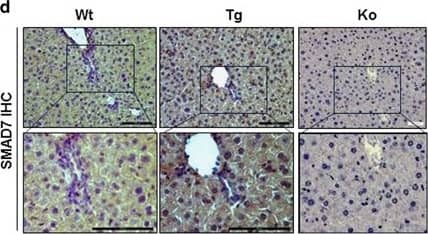
DEN-dependent HCC in Tamoxifen (TAM)-inducible hepatocyte-specific SMAD7 Tg and SMAD7 KO mice. (a) Plasmid constructs used for generating Albumin-SMAD7 Tg and SMAD7fl/fl mice. (b) Experimental design of the DEN-induced HCC in TTR-Cre × SMAD7 Tg or KO mice. (c) Smad7 levels were analyzed by RT–PCR and (d) immunohistochemistry (positive staining is indicated by brown color). Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/oncsis201685), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse Smad7 by Western Blot
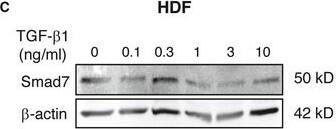
TGF‐ beta1 signaling in MSCs is regulated by environmental TGF‐ beta1 concentrationAAT‐MSCs were cultured in the labeling media containing [35S]‐Met/Cys and increasing concentrations of r.h. TGF‐ beta1 at 0, 0.1, 0.3, 1, 3, and 10 ng/ml for overnight. Supernatants were harvested and incubated with TGF‐ beta1 capture antibody. After washing, the captured TGF‐ beta1 was transferred to glass filter membrane, and the radioactivity was counted with a scintillation counter (black bars). The supernatants were incubated with new TGF‐ beta1 capture antibody, and the radioactivity of captured TGF‐ beta1 was counted again (white bars). The data are given as mean ± SD of five repeated measurements. This experiment was performed four times with comparable results. CPM, count per minute.B, CAT‐MSCs (B) or HDFs (C) were cultured in media containing increasing concentrations of r.h. TGF‐ beta1 at 0, 0.1, 0.3, 1, 3, and 10 ng/ml for 24 h, before protein lysates or total RNAs were harvested. Western blot analysis of Smad7 was performed with protein lysates and semi‐quantitatively analyzed with densitometry. beta‐actin served as loading control. Data are given as mean ± SEM of three independent blots. **P < 0.01, by one‐way ANOVA with Tukey's test. AU, arbitrary unit.DFrom total RNA, the mature miR‐21 and endogenous control RNU6B were converted to cDNA and quantified by qPCR‐based TaqMan microRNA assays. The expression of miR‐21 was normalized on RNU6B. Data are given as mean ± SEM of three independent experiments.Source data are available online for this figure. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32080965), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human Smad7 by Western Blot
TGF‐ beta1 signaling in MSCs is regulated by environmental TGF‐ beta1 concentrationAAT‐MSCs were cultured in the labeling media containing [35S]‐Met/Cys and increasing concentrations of r.h. TGF‐ beta1 at 0, 0.1, 0.3, 1, 3, and 10 ng/ml for overnight. Supernatants were harvested and incubated with TGF‐ beta1 capture antibody. After washing, the captured TGF‐ beta1 was transferred to glass filter membrane, and the radioactivity was counted with a scintillation counter (black bars). The supernatants were incubated with new TGF‐ beta1 capture antibody, and the radioactivity of captured TGF‐ beta1 was counted again (white bars). The data are given as mean ± SD of five repeated measurements. This experiment was performed four times with comparable results. CPM, count per minute.B, CAT‐MSCs (B) or HDFs (C) were cultured in media containing increasing concentrations of r.h. TGF‐ beta1 at 0, 0.1, 0.3, 1, 3, and 10 ng/ml for 24 h, before protein lysates or total RNAs were harvested. Western blot analysis of Smad7 was performed with protein lysates and semi‐quantitatively analyzed with densitometry. beta‐actin served as loading control. Data are given as mean ± SEM of three independent blots. **P < 0.01, by one‐way ANOVA with Tukey's test. AU, arbitrary unit.DFrom total RNA, the mature miR‐21 and endogenous control RNU6B were converted to cDNA and quantified by qPCR‐based TaqMan microRNA assays. The expression of miR‐21 was normalized on RNU6B. Data are given as mean ± SEM of three independent experiments.Source data are available online for this figure. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32080965), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human Smad7 by Western Blot
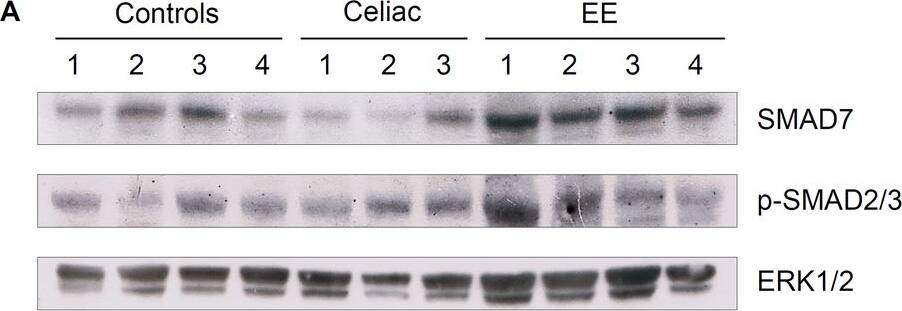
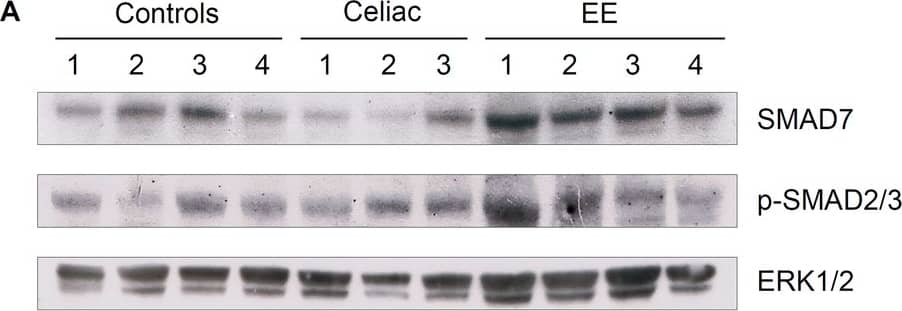
SMAD protein expression data from healthy controls, celiac disease, and environmental enteropathy.A) Representative western blot example investigating SMAD7; p-SMAD2,3 expression (using ERK1/2 as a normalizing protein) from controls (Ctl), celiac disease (CD), and environmental enteropathy (EE) patients. Note that for p-SMAD2,3, patients 3 and 4 have a double band that controls lacked. This band was likely an artifact and consequently was not considered in the densitometric analyses. B) Elevated SMAD7 densitometry levels in EE compared to controls (Ctl mean±SD = 0.47±0.20 a.u., EE = 1.13±0.25 a.u., p-value<0.05). Corresponds to WB5 (see S3 Table). C) Elevated p-SMAD2,3 densitometry levels in EE compared to controls (Ctl mean±SD = 0.38±0.14 a.u., EE = 0.60±0.10 a.u., p-value<0.05). Corresponds to WB2 (see S3 Table). D) Elevated p-SMAD2,3 densitometry levels in EE and CD compared to controls, with significant differences only between EE and controls (p-value<0.05) (Ctl mean±SD = 0.34±0.12 a.u., CD = 0.87±0.36 a.u., EE = 0.97±0.11 a.u.). Corresponds to WB1 (see S3 Table). Image collected and cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pntd.0006224), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human Smad7 by Knockdown Validated
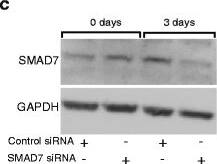
SMAD7 is involved in the regulation of IL-1R1 expression in PSCs. a Gene expression of SMAD7 and IL-1R1 from PSCs established from different patients (n = 19) were analysed by qPCR. PSCs were transfected with non-signaling siRNA (control) or SMAD7 siRNA (100nM) for 3 days. Correlation was determined using Pearson correlation coefficient. *p < 0.05. b Pancreatic stellate cells were incubated in the presence of increasing concentrations of recombinant TGF beta (30 pg/ml – 8 ng/ml) to assess the optimal concentration required to down-regulate the gene expression of IL-1R1 and to investigate the impact of TGF beta on PSC expression of SMAD7. c SMAD7 protein expression was visualized by western blotting after 3 days of culture and (d) IL-1R1 was visualized after additional 2 days in serum free medium Image collected and cropped by CiteAb from the following publication (https://jeccr.biomedcentral.com/articles/10.1186/s13046-016-0400-5), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human Human/Mouse/Rat Smad7 Antibody by Western Blot
SMAD protein expression data from healthy controls, celiac disease, and environmental enteropathy.A) Representative western blot example investigating SMAD7; p-SMAD2,3 expression (using ERK1/2 as a normalizing protein) from controls (Ctl), celiac disease (CD), and environmental enteropathy (EE) patients. Note that for p-SMAD2,3, patients 3 and 4 have a double band that controls lacked. This band was likely an artifact and consequently was not considered in the densitometric analyses. B) Elevated SMAD7 densitometry levels in EE compared to controls (Ctl mean±SD = 0.47±0.20 a.u., EE = 1.13±0.25 a.u., p-value<0.05). Corresponds to WB5 (see S3 Table). C) Elevated p-SMAD2,3 densitometry levels in EE compared to controls (Ctl mean±SD = 0.38±0.14 a.u., EE = 0.60±0.10 a.u., p-value<0.05). Corresponds to WB2 (see S3 Table). D) Elevated p-SMAD2,3 densitometry levels in EE and CD compared to controls, with significant differences only between EE and controls (p-value<0.05) (Ctl mean±SD = 0.34±0.12 a.u., CD = 0.87±0.36 a.u., EE = 0.97±0.11 a.u.). Corresponds to WB1 (see S3 Table). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29415065), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human Human/Mouse/Rat Smad7 Antibody by Western Blot
Adenovirus-mediated SMAD7 expression inhibits STAT3 phosphorylation in HuH-7. (a) Ectopic SMAD7 expression inhibits both endogenous and IL-6-induced STAT3 phosphorylation in HuH-7 cells. (b) Ectopic SMAD7 expression inhibits IL-6-induced MCL-1 expression in HuH-7 cells. Quantification of immunoblot analysis was performed of two independent experiments. One result is shown representatively. (c) IL-6 expression levels were investigated by RT–PCR upon ectopic SMAD7 expression. (d) Theoretical pathway connections between SMAD7 and STAT3 in HCC analyzed by cBioPortal analysis (http://www.cbioportal.org/). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28134936), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Smad7 by Western Blot
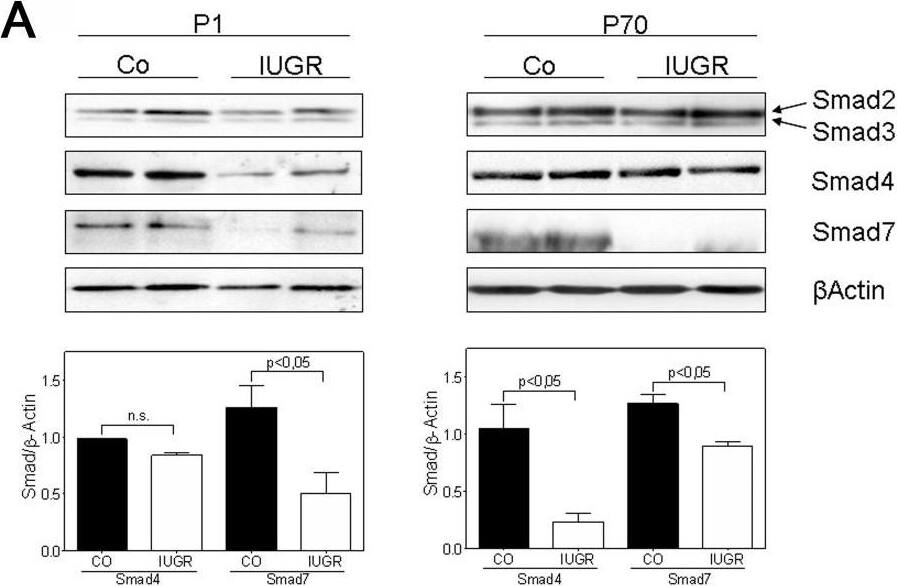
IUGR alters expression and phosphorylation of Smad proteins in rats.A: Representative immunoblots illustrating the expression of TGF-beta -specific Smad2, Smad3, the co-Smad, Smad4, and the inhibitory Smad, Smad7, in lungs extracted at days P1 and P70 from rats with and without IUGR. beta-actin served as loading control. Immunoblot data were quantified for Smad4 and Smad7 for both days P1 and P70 (Co as black bar, and IUGR as white bar); n = 4–6 for each bar. The significance for each bar is indicated by p values, IUGR vs. CO; two-tailed Mann-Whitney test. B: The expression of active TGF-beta signaling components in lung homogenates of rats with and without IUGR was analyzed by immunoblotting of phosphorylated (p) and total Smad2 and Smad3. beta-actin served as loading control. Immunoblot data were quantified for pSmad2 and pSmad3 for both days P1 and P70 (Co as black bar, and IUGR as white bar); n = 4–6 for each bar. The significance for each bar is indicated by p values, IUGR vs. CO; two-tailed Mann-Whitney test. C: Immunhistochemical localization and expression pattern of pSmad2 and pSmad3 in lungs of rats with IUGR (right column) and without IUGR (left column). A–F: representative fields illustrating the expression and localization of pSmad2 in bronchi (A–D) and in the alveoli (E–F) of lungs extracted on day P70. G–L: representative fields illustrating the expression and localization of pSmad3 in bronchi (G–J) and in the alveoli (K–L) of lungs extracted on day P70. M–N: negative control. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/22028866), licensed under a CC-BY license. Not internally tested by R&D Systems.