Cell Proliferation Induced by PDGF‑AA and Neutralization by Mouse PDGF R alpha Antibody.
Recombinant Human PDGF-AA (
221-AA) stimulates proliferation in the NR6R-3T3 mouse fibroblast cell line in a dose-dependent manner (orange line), as measured by Resazurin (
AR002. Proliferation elicited by Recombinant Human PDGF-AA (250 ng/mL) is neutralized (green line) by increasing concentrations of Goat Anti-Mouse PDGF Ra Antigen Affinity-purified Polyclonal Antibody (Catalog # AF1062). The ND
50 is typically 0.2-1.6 µg/mL.
Detection of Rat PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165017.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165013.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-21202416507.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165015.jpg
Detection of Mouse PDGF R alpha by Immunohistochemistry.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165011.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165018.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165010.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165014.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-212024165012.jpg
Detection of Mouse PDGF R alpha by Immunocytochemistry/Immunofluorescence.
af1062_mouse-pdgf-r-alpha-affinity-purified-polyclonal-ab-21202416509.jpg
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha driven brain tumors display features of high grade glioma.(a–g) Histopathological analysis of tumor areas by H&E staining shows a high concentration of mitotic figures (a, arrows), high cellularity and nuclear atypia (b), perineuronal satellitosis (c; N, neuronal nuclei), perivascular growth (d), intrafascicular growth (e), subarachnoid spreading (f), and areas of incipient necrosis (g; arrows point to pyknotic nuclei). (h–k) IF labeling of brain tumor sections for cell type specific markers. Nuclei labeled with DAPI are shown in blue. Tumor cells with high PDGFR alpha expression were highly proliferative, as seen by proliferation marker Ki67 (h), and express the OPC cell lineage markers Olig2, Sox2, Sox10, and Ng2, as well as the neural stem cell marker Nestin (i–k). Tumor cells were negative for immunosignal of astroglial marker GFAP, mature oligodendrocyte marker APC-CC1, and neuronal marker NeuN (l–n). Scale bars: 10 μm (a–g), 20 μm (h–n). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25683249), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha driven brain tumors display features of high grade glioma.(a–g) Histopathological analysis of tumor areas by H&E staining shows a high concentration of mitotic figures (a, arrows), high cellularity and nuclear atypia (b), perineuronal satellitosis (c; N, neuronal nuclei), perivascular growth (d), intrafascicular growth (e), subarachnoid spreading (f), and areas of incipient necrosis (g; arrows point to pyknotic nuclei). (h–k) IF labeling of brain tumor sections for cell type specific markers. Nuclei labeled with DAPI are shown in blue. Tumor cells with high PDGFR alpha expression were highly proliferative, as seen by proliferation marker Ki67 (h), and express the OPC cell lineage markers Olig2, Sox2, Sox10, and Ng2, as well as the neural stem cell marker Nestin (i–k). Tumor cells were negative for immunosignal of astroglial marker GFAP, mature oligodendrocyte marker APC-CC1, and neuronal marker NeuN (l–n). Scale bars: 10 μm (a–g), 20 μm (h–n). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25683249), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha driven brain tumor model.(a) Schematic diagram of PDGFR alpha J/K knock-in alleles. ATG, start codon; SA, splice acceptor; STOP, PGK-neo cassette. (b) Kaplan-Meier survival curves of 4 mouse mutant cohorts with brain tumors. Mice generally succumbed to subcutaneous fibrosarcomas, and brain tumors were detected by histological analysis. (c) Example of early stage tumor growth, as revealed by IHC for proliferation marker Ki67 and PDGFR alpha. Note high density of Ki67+ proliferating cells in tumor area, increased expression level of PDGFR alpha, and invasive migration of tumor cells through corpus callosum into contralateral hemisphere. (d) H&E staining of an advanced brain tumor growth (asterisk in tumor centre, dashed line demarcates expansion). Scale bars: 50 μm (c, d). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25683249), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
Etv5 is necessary and sufficient for proliferation and cell fate bias downstream of Cic loss. a–f Cic-null NSCs (CicnullEmpty), Cic-null NSCs with dominant negative Etv5 (CicnullDN-Etv5), and Cic-wildtype NSCs overexpressing Etv5 (Etv5 overpression) were grown in lineage-directed culture conditions and assessed for their ability to differentiate to neurons, astrocytes, and oligodendrocytes as determined by immunostaining for bIII-Tubulin (Tuj1), Gfap, and Olig2, Pdgfra, and Mbp. Scale bar: 10 μm. Analysis of Tuj1 + cells from NSCs in neuronal differentiating condition, NDC a, d; analysis of Gfap + cells from NSCs in astrocytic differentiating condition, ADC b, e; and analyses of Olig2 + , Pdgfra + , and Mbp + cells from NSCs in oligodendrocyte differentiating condition, ODC c, f from n = 3 biological replicates, with three technical replicates each, for cell culture studies. g, h Representative images and quantitation of EdU incorporation 2 days post electroporation of wildtype ETV5 or empty control plasmid, both carrying mCherry as a marker, into E13 CICFl/Fl VZ. Note: mCherry fluorescence and EdU staining were false-colorized to green and red after grayscale imaging. Scale bar: 50 μm. Data from n = 4 mice per each group. Scale bar: 50 μm. i, j Representative images and quantitation of EdU incorporation 2 days post-electroporation of Cre only or of Cre co-electroporated with DNETV5 into E13 CICFl/Fl VZ. Data from n = 4 mice per each group. Scale bar: 50 μm k EdU incorporation assay in cultured Cic-wildtype NSCs without or with ETV5 overexpression from n ≥ 3 biological replicates. l EdU incorporation in Cic-floxed NSCs with Cre, and without or with DNETV5 expression from n ≥ 3 biological replicates. Data shown as mean ± SD. Statistical analyses performed either t test in h, j, k, l; or with ANOVA with Tukey’s post hoc test in d, e, f. ns–not significant, *p < 0.05, **p < 0.01, ***p < 0.0001. Source data are provided as a Source Data file. ADC–astrocytic differentiation condition, NDC–neuronal differentiation condition, ODC–oligodendrocytic differentiation condition. VZ–ventricular zone, LV–lateral ventricle Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31043608), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
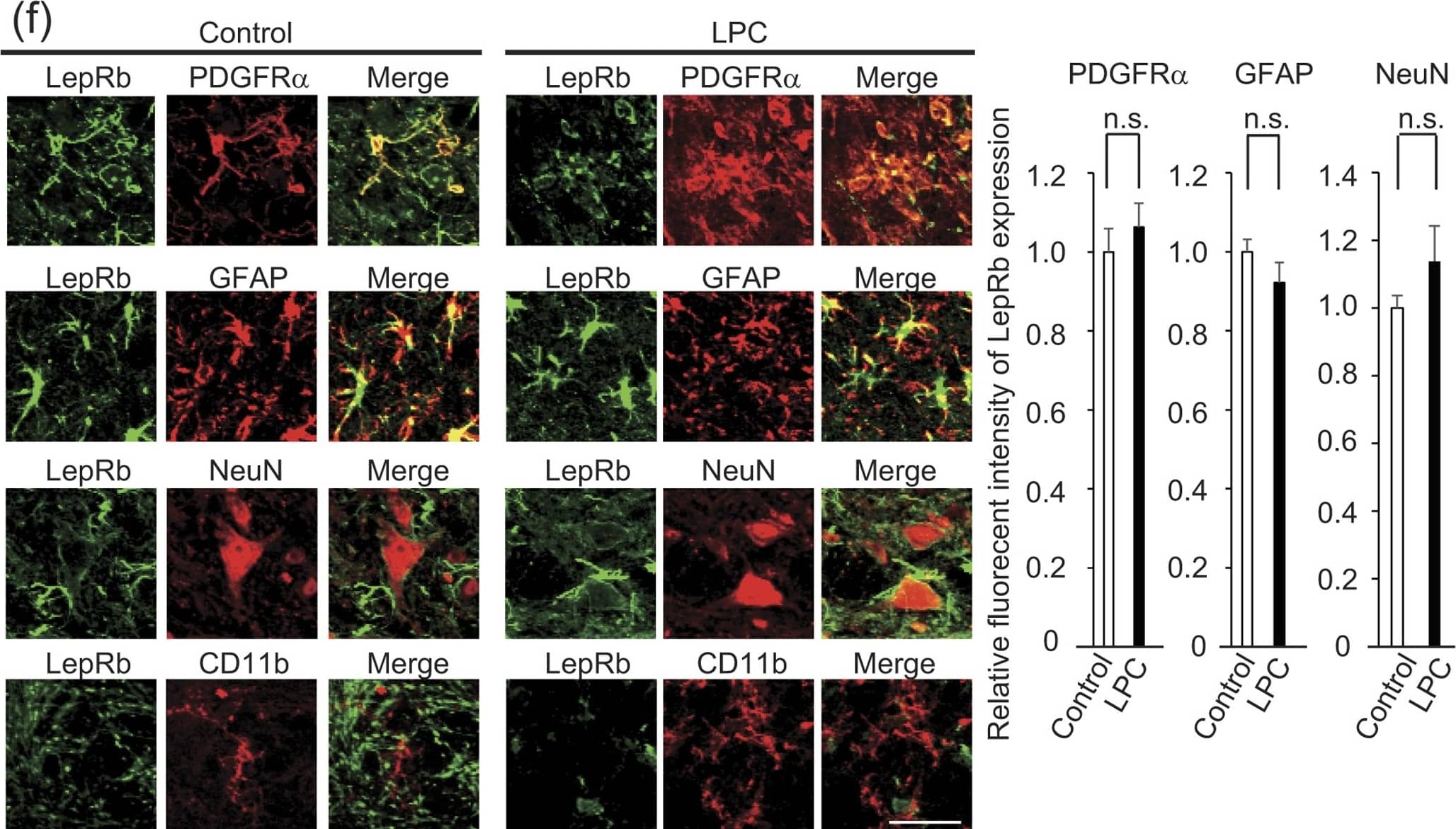
LPC injection does not enhance leptin expression in the CNS.(a) Representative images of MBP expression in a mouse spinal cord 14 days after LPC injection are shown; the graph shows quantification of the demyelinating area in the dorsal spinal cord (n = 3–4). P = 0.001542, Student’s t-test. (b) Representative images of NeuN expression in a mouse spinal cord 14 days after LPC injection; the graph shows quantification of the density of NeuN-positive cells in the spinal cord (n = 3). P = 0.299940, Student’s t-test, n.s. indicates no significant difference. (c) Quantification of leptin protein expression in indicated organs. Tissues were obtained from the mice 3 days after LPC injection (n = 3 for control, 4 for LPC injection). P = 0.318966 (adipose tissue), 0.10446 (brain stem), 0.332281 (cerebellum), 0.345245 (liver), 0.453104 (kidney), 0.098135 (heart), 0.335722 (lung), 0.236771 (muscle), 0.44662 (spleen), 0.465966 (stomach). Student’s t-test. n.s. indicates no significant difference. (d) Quantification of spinal cord leptin protein 3 days after LPC injection (n = 3 for control, 4 for LPC injection). P = 0.026865, Student’s t-test. (e) Quantification of spinal cord leptin mRNA 3 days after LPC injection (n = 6). P = 0.324930, Student’s t-test, n.s. indicates no significant difference. (f) Representative images of LepRb (green) expression in combination with PDGFR alpha, GFAP NeuN, and CD11b (red) in the mouse spinal cord with or without LPC injection. Spinal cord sections were obtained 3 days after LPC injection. Graph indicates the relative intensity of leptin protein expression in indicated cell type (n = 3). P = 0.287452 (PDGFR alpha), 0.181059 (GFAP), 0.199972 (NeuN), Student’s t-test, n.s. indicates no significant difference. *P < 0.05, **P < 0.01, error bars represent SEM. Scale bars; 100 μm for (a and b), 25 μm for (f). Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/srep40397), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
Leptin promotes OPC proliferation.(a) Representative image of cultured OPCs stained with antibodies against LepRb (green) and PDGFR alpha (red). Scale bar: 25 μm. (b) Relative BrdU incorporation into the OPC obtained from the brain (left graph) and spinal cord (right graph). Cells were treated with recombinant leptin for 48 h (n = 4). (Left graph) P = 0.005993 (control vs 10 ng/mL), 0.045616 (control vs 100 ng/mL), (Right graph) P = 0.004456 (control vs 10 ng/mL), 0.017859 (control vs 100 ng/mL). (c) Relative BrdU incorporation into the OPC after leptin stimulation (10 ng/ml) with U0126 (20 μM), a MEK inhibitor (n = 4 for brain OPCs, n = 3 for spinal cord OPCs). (Left graph) P = 0.019753 (control vs leptin), 0.039433 (leptin vs leptin + U0126), (Right graph) P = 0.045545 (control vs leptin), 0.04486 (leptin vs leptin + U0126). (d) Representative images of western blotting (upper panels) and quantitative analysis of ERK phosphorylation (lower graph) are shown. OPCs were treated with leptin (10 ng/ml) under indicated periods (n = 3). P = 0.006352 (2 min), 0.016571 (5 min), 0.017675 (10 min), 0.024100 (15 min), 0.081342 (30 min). *P < 0.05, **P < 0.01, ANOVA with Tukey’s post-hoc test. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/srep40397), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Rat PDGFR alpha by Immunocytochemistry/Immunofluorescence
Extracellularly applied recombinant human alpha-syn PFFs induced cytoplasmic alpha-syn-immunoreactive inclusions in primary BCAS1(+) cell cultures. a Confocal images of BCAS1(+) cells, which were incubated with 1 μM alpha-syn PFFs for 24 h from day 4 after differentiation induction, showing the intracellular inclusions labeled with both anti-alpha -syn antibody and thioflavin S. Scale bar = 5 μm. b Immunostaining of oligodendroglial cells incubated with 1 μM alpha-syn PFFs for 24 h from days 3 (upper) and 4 (lower) after differentiation induction showing the ubiquitous development of thioflavin S-labeled inclusions in PDGFR alpha(+) cells and BCAS1(+) cells. In contrast, few BCAS1(−)/MBP(+) cells developed thioflavin S-labeled inclusions. Scale bar = 50 μm. c The percentages of oligodendroglial cells containing thioflavin S-labeled inclusions were compared between BCAS1(−)/PDGFR alpha(+) cells and BCAS1(+)/PDGFR alpha(+) cells (upper, performed on day 3), and between BCAS1(+)/MBP(+) cells and BCAS1(−)/MBP(+) cells (lower, performed on day 4). N = 4, respectively, independent culture, Mann–Whitney, p* < 0.05 Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32727582), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
Transplantation of CD11b/LIF transgenic BMCs reduces the numbers of FAPs in dystrophic muscle but does not affect phenotype. a QPCR analysis shows that TA muscles from LIF BMT/mdx recipients have reduced Pdgfra gene expression. N = 7 or 8 for WT BMT/mdx and LIF BMT/mdx data sets, respectively, * indicates significantly different from WT BMT/mdx recipients at P < 0.05. P-values based on two-tailed t-test. For all histograms in the figure, the bars indicate mean ± sem. b To quantify the number of FAPs, muscle sections were co-labeled with antibodies to PDGFR alpha (red) and CD31, CD45 (green). Arrowheads indicate FAPs (CD31-CD45-PDGFR alpha+). Bar = 50 μm. c Fewer FAPs (CD31-CD45-PDGFR alpha+) in TA cross-sections of LIF BMT/mdx recipients compared to WT BMT/mdx recipients. N = 5 for each data set. d There was no detectible change in phenotype of PDGFR alpha+ cells assayed for co-expression of the fibrogenic marker HSP47. e FACS plots demonstrating strategy for sorting FAPs (Hoechst + CD11b-CD31-CD45-PDGFR alpha+). Fibroblasts derived from FAPs were stimulated with LIF (10 ng/ml) and/or TGF beta1 (10 ng/ml) for 3 h (f–h) or 3 days (i–k) and assayed by QPCR for Ctgf (f, i), Fn1 (g, j), and Col1a1 (h, k). N = 4 technical replicates for each data set. Significant findings were verified with biological replicates of cells sorted from independent donors. * Indicates significantly different from control cultures, # indicates significantly different from TGF beta1 treated cultures, and Φ indicates significantly different from LIF-treated cultures at P < 0.05. P-values based on ANOVA with Tukey’s multiple comparison test. Source data are provided as a Source Data file Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31243277), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
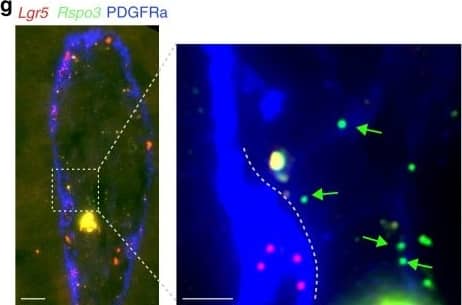
Lgr5 is expressed abundantly in villus tip telocytes.a smFISH of Lgr5, DAPI in blue, Scale bar–20 µm. b Blow up of villus tip, Scale bar–10 µm. In a, b thin white arrows point at autofluorescent blobs. c blow up of crypt, Scale bar–10 µm. Red arrows in b–c point to Lgr5 positive cells. dLgr5 mRNA (red dots) expressed in PDGFRa+ VTTs that co-express Bmp4 mRNA (green dots). Scale bar–10 µm. Red arrows point to Lgr5 and Bmp4 double positive cells. e Blow up of the region boxed in d. Scale bar–5 µm. fLgr5 mRNA concentrations in VTTs are comparable to those in Lgr5+ crypt base columnar cells (n = 25 cells examined over 2 mice for each region). Boxes show 25–75 percentiles of the smFISH expression, horizontal red lines are medians. Whiskers, extend to the most extreme data point within 1.5× the interquartile range (IQR) from the box; g) Rspo3 mRNAs are localized on telopodes that extend away from the cell bodies of the VTTs. VTTs are marked by Lgr5 mRNA (red dots), Rspo3 mRNA (green dots) is localized away from the cell body, PDGFRa antibody mark VTTs cell bodies and telopodes. Scale bar–10 µm, in inset, green arrows point to Rspo3 mRNAs (green dots) localized on PDGFRa telopodes (blue). Telocyte cell body is marked by white dashed line. inset Scale bar–5 µm. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32321913), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha driven brain tumors display features of high grade glioma.(a–g) Histopathological analysis of tumor areas by H&E staining shows a high concentration of mitotic figures (a, arrows), high cellularity and nuclear atypia (b), perineuronal satellitosis (c; N, neuronal nuclei), perivascular growth (d), intrafascicular growth (e), subarachnoid spreading (f), and areas of incipient necrosis (g; arrows point to pyknotic nuclei). (h–k) IF labeling of brain tumor sections for cell type specific markers. Nuclei labeled with DAPI are shown in blue. Tumor cells with high PDGFR alpha expression were highly proliferative, as seen by proliferation marker Ki67 (h), and express the OPC cell lineage markers Olig2, Sox2, Sox10, and Ng2, as well as the neural stem cell marker Nestin (i–k). Tumor cells were negative for immunosignal of astroglial marker GFAP, mature oligodendrocyte marker APC-CC1, and neuronal marker NeuN (l–n). Scale bars: 10 μm (a–g), 20 μm (h–n). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25683249), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
Lgr5 is expressed abundantly in villus tip telocytes.a smFISH of Lgr5, DAPI in blue, Scale bar–20 µm. b Blow up of villus tip, Scale bar–10 µm. In a, b thin white arrows point at autofluorescent blobs. c blow up of crypt, Scale bar–10 µm. Red arrows in b–c point to Lgr5 positive cells. dLgr5 mRNA (red dots) expressed in PDGFRa+ VTTs that co-express Bmp4 mRNA (green dots). Scale bar–10 µm. Red arrows point to Lgr5 and Bmp4 double positive cells. e Blow up of the region boxed in d. Scale bar–5 µm. fLgr5 mRNA concentrations in VTTs are comparable to those in Lgr5+ crypt base columnar cells (n = 25 cells examined over 2 mice for each region). Boxes show 25–75 percentiles of the smFISH expression, horizontal red lines are medians. Whiskers, extend to the most extreme data point within 1.5× the interquartile range (IQR) from the box; g) Rspo3 mRNAs are localized on telopodes that extend away from the cell bodies of the VTTs. VTTs are marked by Lgr5 mRNA (red dots), Rspo3 mRNA (green dots) is localized away from the cell body, PDGFRa antibody mark VTTs cell bodies and telopodes. Scale bar–10 µm, in inset, green arrows point to Rspo3 mRNAs (green dots) localized on PDGFRa telopodes (blue). Telocyte cell body is marked by white dashed line. inset Scale bar–5 µm. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32321913), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
OPC expresses leptin receptors.(a) Representative images of spinal cord sections, which were double-labeled for LepRb (green) in combination with PDGFR alpha (red). Spinal cord sections were obtained 7 days after LPC injection; the graph shows quantification (n = 3). P = 0.007573 (LepRb flox vs intact CKO), 0.0108779 (LepRb flox vs LPC CKO), ANOVA with Tukey’s post-hoc test. (b) Relative expression of leptin receptors mRNA in PDGFR alpha-positive OPC obtained from the brain of PDGFR alpha-creERT:: Lepr flox/flox mice and +/+::Lepr flox/flox mice (n = 5,6). P = 0.005878 (LepRa), 0.010306 (LepRb), 0.001535 (LepRc), 0.003169 (LepRd), 0.030459 (LepRe), Student’s t-test. (c) Representative images of spinal cord sections which were double labeled for BrdU in combination with PDGFR alpha (left panels) and GSTπ (right panels). Sections were prepared 7 days (left panels) and 14 days (right panels) after LPC injection. BrdU was administrated during 3–7 days after LPC injection; the graph shows quantification (n = 5–8). P = 0.029791(PDGFR alpha and BrdU labeled cells), 0.028870 (GSTπ and BrdU labeled cells), Student’s t-test. (d) Representative images of PDGFR alpha expression in the intact spinal cord of PDGFR alpha-creERT:: Lepr flox/flox mice and +/+::Lepr flox/flox mice; the graph shows quantification (n = 3–4). P = 0.404999, Student’s t-test, n.s. indicates no significant difference. (e) Representative images of APC expression in the intact spinal cord of PDGFR alpha-creERT:: Lepr flox/flox mice and +/+::Lepr flox/flox mice; the graph shows quantification (n = 3). P = 0.495667, Student’s t-test, n.s. indicates no significant difference. (f) Representative spinal cord section of PDGFR alpha-creERT:: Lepr flox/flox mice, which were prepared 14 days after LPC injection and stained with MBP; the graph shows quantification of the demyelinating area in the dorsal spinal cord (n = 7 for control, 10 for CKO). P = 0.030688, Student’s t-test. (g) Representative spinal cord sections which were labeled for CD11b. Sections were prepared 7 days after LPC injection. The graph shows quantification (n = 3). P = 0.493264, Student’s t-test, n.s. indicates no significant difference. *P < 0.05, **P < 0.01, error bars represent SEM. Scale bars: 25 μm for (a), 50 μm for high magnification images in (c), 100 μm for others. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/srep40397), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunocytochemistry/Immunofluorescence
Endogenous leptin sustains spontaneous OPC proliferation.(a) Representative images of spinal cord sections, which were prepared 7 days (left panels) and 14 days (right panels) after LPC injection and double labeled for BrdU in combination with PDGFR alpha (upper panels), GSTπ (upper panels) and olig2 (lower panels). BrdU was administrated during 3–7 days after LPC injection; the graph shows quantification (n = 5–8). P = 0.042915 (PDGFR alpha and BrdU labeled cells), 0.013560 (Olig2 and BrdU labeled cells 7 days after injection), 0.012111 (GSTπ and BrdU labeled cells), 0.009797 (Olig2 and BrdU labeled cells 14 days after injection), Student’s t-test. (b) Representative spinal cord sections, which were prepared 14 days after LPC injection and stained with MBP are shown; the graph shows quantification (n = 6). P = 0.025243, Student’s t-test. (c) Representative images of spinal cord section, which were prepared 7 days after LPC injection and labeled for CD11b; the graph shows quantification (n = 4). P = 0.213763, Student’s t-test, n.s. indicates no significant difference. *P < 0.05, **P < 0.01, error bars represent SEM. Scale bar: 50 μm for high magnification images in a, 100 μm for others. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/srep40397), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha driven brain tumors display features of high grade glioma.(a–g) Histopathological analysis of tumor areas by H&E staining shows a high concentration of mitotic figures (a, arrows), high cellularity and nuclear atypia (b), perineuronal satellitosis (c; N, neuronal nuclei), perivascular growth (d), intrafascicular growth (e), subarachnoid spreading (f), and areas of incipient necrosis (g; arrows point to pyknotic nuclei). (h–k) IF labeling of brain tumor sections for cell type specific markers. Nuclei labeled with DAPI are shown in blue. Tumor cells with high PDGFR alpha expression were highly proliferative, as seen by proliferation marker Ki67 (h), and express the OPC cell lineage markers Olig2, Sox2, Sox10, and Ng2, as well as the neural stem cell marker Nestin (i–k). Tumor cells were negative for immunosignal of astroglial marker GFAP, mature oligodendrocyte marker APC-CC1, and neuronal marker NeuN (l–n). Scale bars: 10 μm (a–g), 20 μm (h–n). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25683249), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha driven brain tumors display features of high grade glioma.(a–g) Histopathological analysis of tumor areas by H&E staining shows a high concentration of mitotic figures (a, arrows), high cellularity and nuclear atypia (b), perineuronal satellitosis (c; N, neuronal nuclei), perivascular growth (d), intrafascicular growth (e), subarachnoid spreading (f), and areas of incipient necrosis (g; arrows point to pyknotic nuclei). (h–k) IF labeling of brain tumor sections for cell type specific markers. Nuclei labeled with DAPI are shown in blue. Tumor cells with high PDGFR alpha expression were highly proliferative, as seen by proliferation marker Ki67 (h), and express the OPC cell lineage markers Olig2, Sox2, Sox10, and Ng2, as well as the neural stem cell marker Nestin (i–k). Tumor cells were negative for immunosignal of astroglial marker GFAP, mature oligodendrocyte marker APC-CC1, and neuronal marker NeuN (l–n). Scale bars: 10 μm (a–g), 20 μm (h–n). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25683249), licensed under a CC-BY license. Not internally tested by R&D Systems.
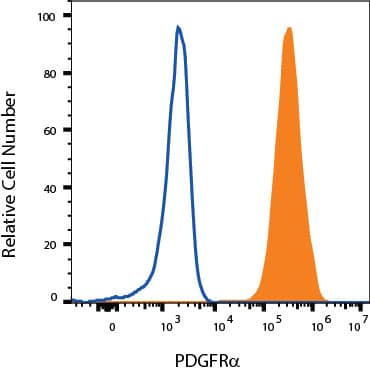
Detection of PDGF R alpha in 3T3-L1 cells by Flow Cytometry
3T3-L1 cells were stained with Goat Anti-Mouse PDGF R alpha Antigen Affinity-purified Polyclonal Antibody (Catalog # AF1062, filled histogram) or isotype control antibody (Catalog # 4-001-A, open histogram) followed by Allophycocyanin-conjugated Anti-Goat IgG Secondary Antibody (Catalog #
F0108). View our protocol for Staining Membrane-associated Proteins.
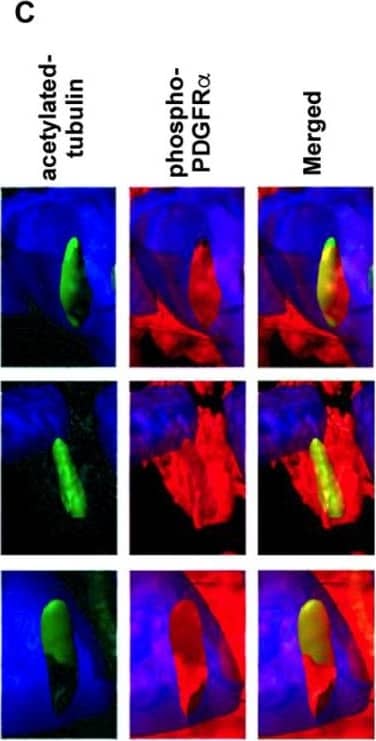
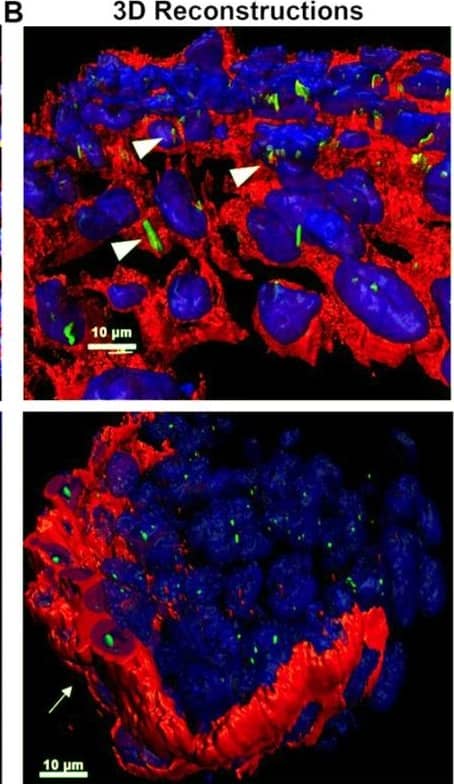
Detection of Mouse PDGFR alpha by Immunohistochemistry
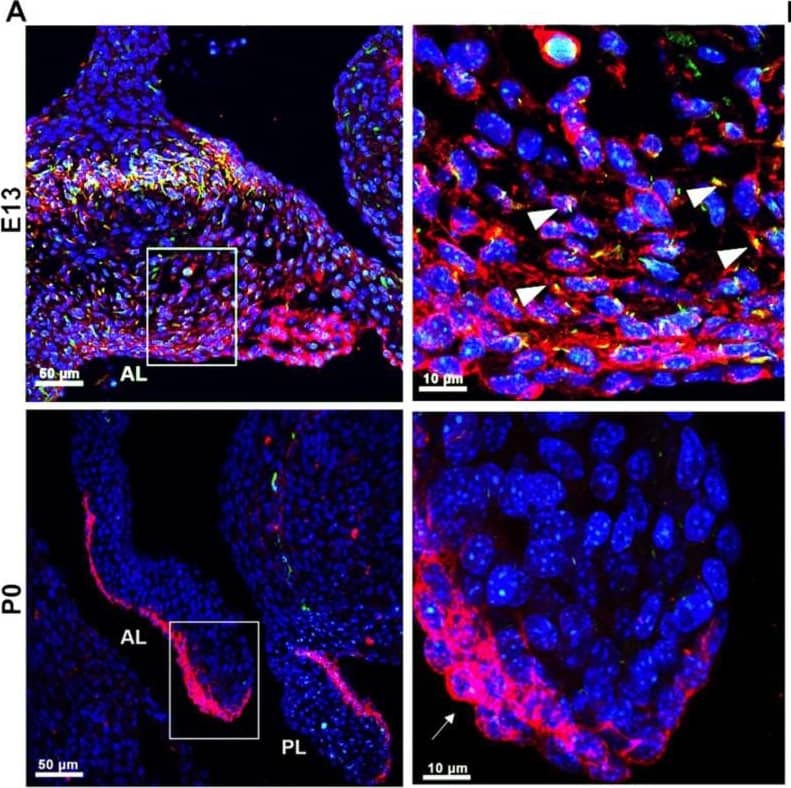
PDGFR alpha and ciliary axonemes early in development. (A) High-resolution microscopy and (B,C) 3D IHC reconstructions of 15 µm wildtype tissue show phosphorylated PDGFR alpha (red) present along ciliary axonemes (acetylated tubulin, green) (arrowheads highlighted in C) at E13. By P0, the receptor localizes to the endocardium where cilia are absent (arrows). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33805717), licensed under a CC-BY license. Not internally tested by R&D Systems.
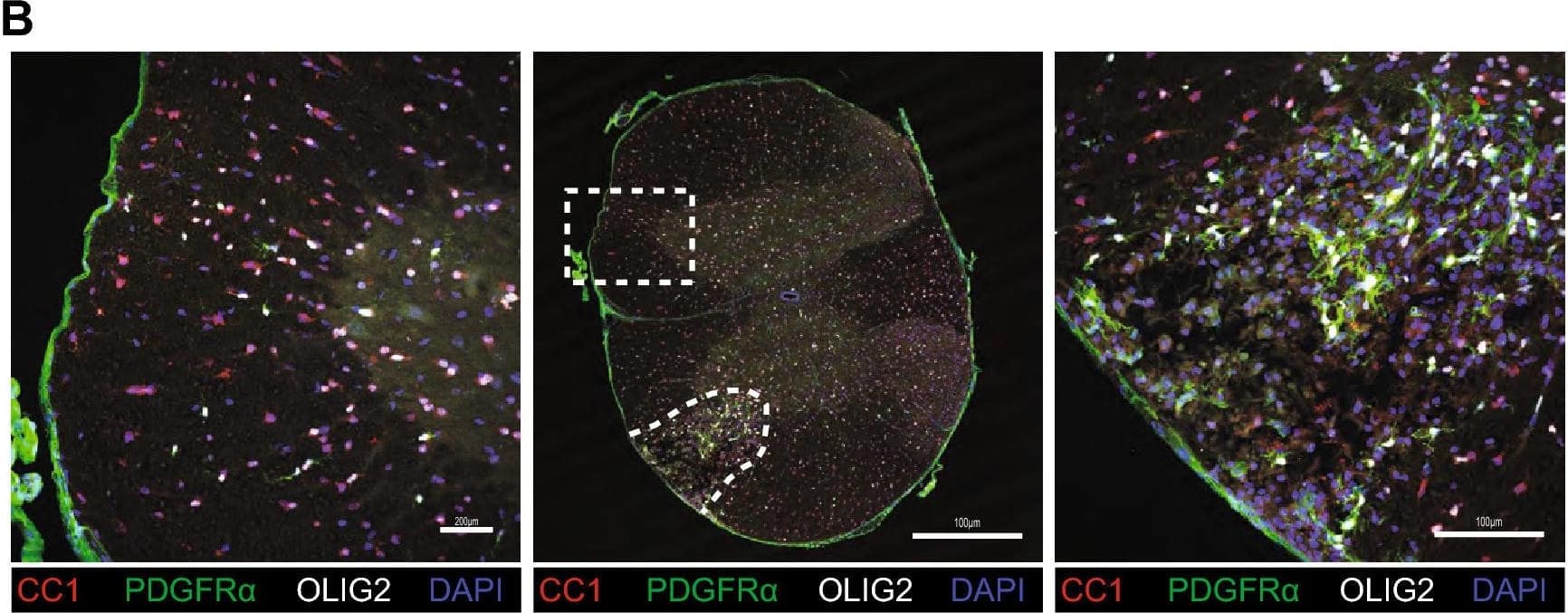
Detection of Mouse PDGFR alpha by Immunohistochemistry
Impact of LPC surgery on mice subjected to running wheel. (A) Workflow and representative eriochrome cyanine-stained sections with lesion in the ventrolateral white matter delineated by dashed lines. Figure was drawn using BioRender. (B) Representative images of LPC lesion (left) and contralateral normal appearing white matter (right) 4 days post injury stained for mature oligodendrocytes (CC1) in red, OPCs (PDGFR alpha) in green, oligodendrocyte lineage cells (OLIG2) in white, and DNA (DAPI) in blue. (C) Representative images of LPC lesion 4 days post injury stained for myelin and myelin debris (MBP) in red, axons (NFH) in green, and astrocytes (GFAP) in white, and DNA (DAPI) in blue. In both (B) and (C), the lesion is outlined by the irregular dashed line while the non-involved contralateral site is denoted by the rectangle dashed line. Scale bar represents 100 μm. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33790323), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha and ciliary axonemes early in development. (A) High-resolution microscopy and (B,C) 3D IHC reconstructions of 15 µm wildtype tissue show phosphorylated PDGFR alpha (red) present along ciliary axonemes (acetylated tubulin, green) (arrowheads highlighted in C) at E13. By P0, the receptor localizes to the endocardium where cilia are absent (arrows). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33805717), licensed under a CC-BY license. Not internally tested by R&D Systems.
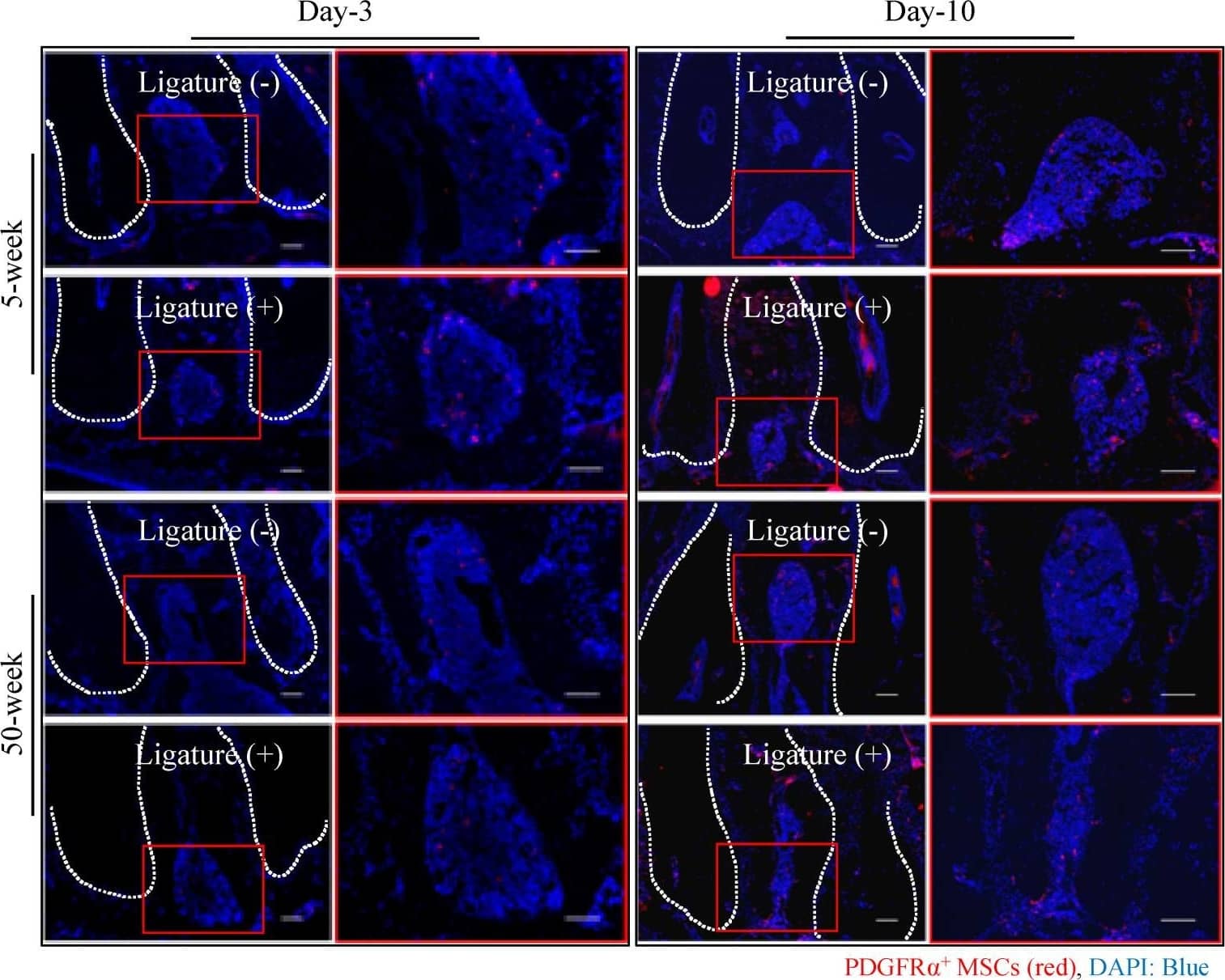
Detection of Mouse PDGFR alpha by Immunohistochemistry
Reduced number of MSCs at the periodontitis site in aged mice. Immunofluorescence images show the number of platelet-derived growth factor receptor alpha (PDGFR alpha)+ MSCs (red) in the furcation area in young and aged mice. Cell nuclei were stained with DAPI (blue). Bar: 100 µm. The graph shows the quantitative analysis indicating that the number of PDGFR alpha+ Mesenchymal stem cells (MSCs) is decreased in aged mice, more prominently at day-10 after ligation. The bar graph represents the mean ± standard deviation of at least three independent samples. * p < 0.05, ** p < 0.01, *** p < 0.001, two-way ANOVA, Tukey test (n = 3). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33143068), licensed under a CC-BY license. Not internally tested by R&D Systems.
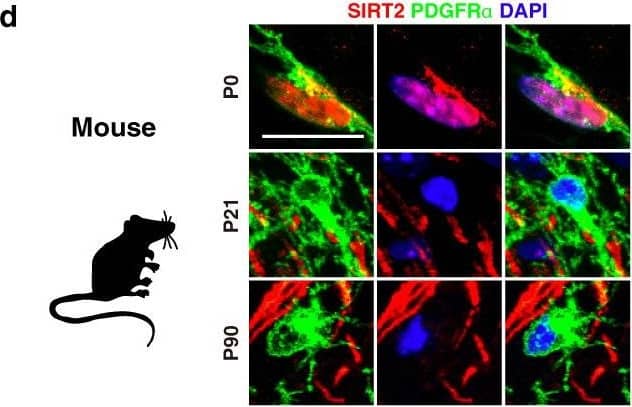
Detection of Mouse PDGFR alpha by Immunohistochemistry
Nuclear entry of SIRT2 in OPCs during remyelination is impaired in the aged mice in vivo.a Primary culture of various types of cells from P0 rat cortex. b qRT-PCR of seven members of sirtuins in primary cultured OPCs and mature oligodendrocytes from the cortex of P0 rat (n = 3). c qRT-PCR of sirt2 in primary cultured various types of cells from the cortex of P0 rat (n = 3). d–f Immunofluorescence and quantification of SIRT2+ cells in the cortex of mice at different ages (n = 3). Scale bar, 10 μm (d), 50 μm (upper panel images of f), 5 μm (lower panel images of f). g–h Immunofluorescence of SIRT2 in the cortex of marmosets at postnatal day 3 (P3, g) or age of 8 years (h). Scale bar, 50 μm. i Immunohistochemistry of SIRT2 in the cortex of human at age of 53 years. Scale bar, 20 μm. j, k Relative SIRT2 protein level in brains of WT young (6 M) and old (18 M) mice (n = 3). l–o Immunofluorescence and quantification of SIRT2+ OPCs and nuclear SIRT2+ OPCs in corpus callosum of WT young and old mice (n = 3). NL, non-lesion, L, demyelination lesion induced by LPC at 5 dpl. Scale bar, 10 μm. All data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed t-test (k) or one-way ANOVA followed by Tukey’s post hoc test (m–o). In all instances ***p < 0.001. n.s. no significance. In (k), **p = 0.004; in (m), **p = 0.002 (L-Young vs. L-Old); in (n), **p = 0.007 (L-Young vs. L-Old). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/35264567), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse PDGFR alpha by Immunohistochemistry
PDGFR alpha and ciliary axonemes early in development. (A) High-resolution microscopy and (B,C) 3D IHC reconstructions of 15 µm wildtype tissue show phosphorylated PDGFR alpha (red) present along ciliary axonemes (acetylated tubulin, green) (arrowheads highlighted in C) at E13. By P0, the receptor localizes to the endocardium where cilia are absent (arrows). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33805717), licensed under a CC-BY license. Not internally tested by R&D Systems.