Mouse TRAIL/TNFSF10 Antibody
R&D Systems, part of Bio-Techne | Catalog # AF1121


Key Product Details
Validated by
Species Reactivity
Validated:
Cited:
Applications
Validated:
Cited:
Label
Antibody Source
Product Specifications
Immunogen
Pro118-Asn291
Accession # P50592
Specificity
Clonality
Host
Isotype
Scientific Data Images for Mouse TRAIL/TNFSF10 Antibody
TRAIL/TNFSF10 in Mouse Thymus.
TRAIL/TNFSF10 was detected in perfusion fixed frozen sections of mouse thymus using Goat Anti-Mouse TRAIL/TNFSF10 Antigen Affinity-purified Polyclonal Antibody (Catalog # AF1121) at 1.5 µg/mL overnight at 4 °C. Tissue was stained using the Anti-Goat HRP-DAB Cell & Tissue Staining Kit (brown; CTS008) and counterstained with hematoxylin (blue). View our protocol for Chromogenic IHC Staining of Frozen Tissue Sections.Detection of Mouse TRAIL/TNFSF10 by Immunohistochemistry
OSCAR–collagen binding co-stimulates IL-1 beta-induced apoptotic signaling in articular chondrocytes.a Caspase activity in mouse articular chondrocytes treated with IL-1 beta (5 ng mL−1) and collagenase (Coll, 50 U mL−1). b, c qRT-PCR analysis (b) and western blotting (c) in mouse articular chondrocytes. Chondrocytes were either untreated or transfected with control siRNA (C-siRNA) (100 nM) or siRNAs specific for mouse Oscar and exposed to IL-1 beta (5 ng mL−1) and collagenase (50 U mL−1) for 48 h. d Articular chondrocyte viability was quantified by MTT assay. Error bars represent mean ± S.E.M. of n = 5 wells. e Caspase-8 and caspase-3 activity was measured using the respective assay kits. f, g qRT-PCR analysis (f) and western blotting (g) in mouse articular chondrocytes. Chondrocytes were treated with hIgG or hOSCAR-Fc (10 μg ml−1) with collagenase and exposed to IL-1 beta for 48 h. h Apoptotic articular chondrocytes were detected and quantified by TUNEL assay. Scale bar = 100 μm. i, j qRT-PCR analysis (i) and western blotting (j) in mouse articular chondrocytes treated with 10 μΜ OSCAR-binding triple-helical peptide before IL-1 beta treatment. Articular chondrocytes were treated with hOSCAR-Fc for 48 h. Error bars shown in a, b, e, f, h and i are mean ± S.E.M. for n = 4 independent experiments. One-way ANOVA was performed followed by Dunnett’s Multiple Comparison’s test (a, i) and Tukey’s Multiple Comparison’s test (b, d, e, f, h), with p values indicated in figure. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32859940), licensed under a CC-BY license. Not internally tested by R&D Systems.Detection of Mouse TRAIL/TNFSF10 by Immunohistochemistry
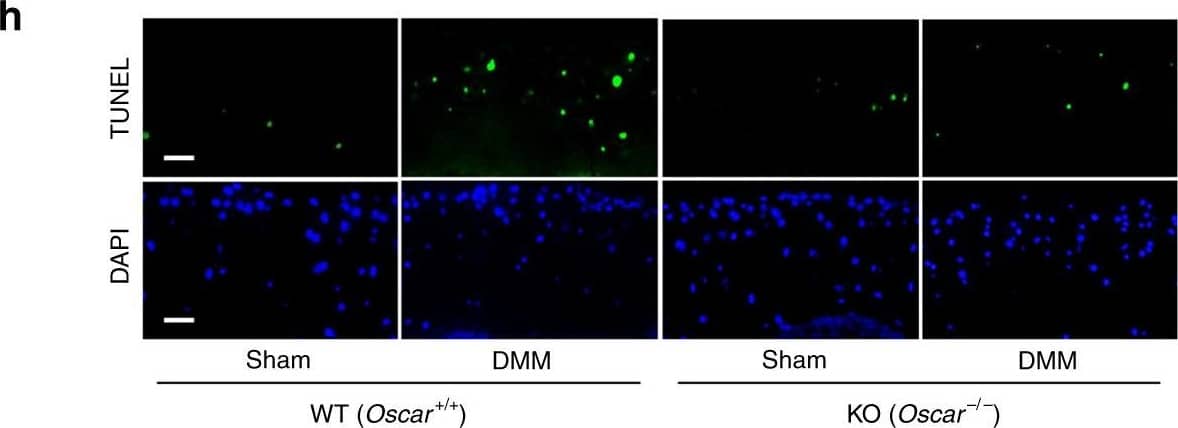
OSCAR regulates OA pathogenesis via TRAIL-induced articular chondrocyte apoptosis.a Selection of putative Oscar targets involved in OA pathogenesis. Venn diagram indicating the number of genes attenuated in articular cartilage Oscar−/− mice after DMM surgery (left). Hypergeometric p value measuring the significance of enrichment is depicted. Gene-wise scaled gene expression patterns of 1270 common genes in each sample are shown (right). b Functional annotations significantly enriched for 1270 common genes. Hypergeometric tests were performed using the hallmark gene annotation in MsigDB, yielding enrichment scores, defined as −log10 (FDR). c Simplified apoptotic signaling pathway altered by Oscar deficiency. Each gene is colored with the DIF score, defined as the extent to which gene expression is altered by Oscar deficiency. A negative DIF indicates transcriptional suppression in Oscar−/− mice. The network was visualized by Cytoscape v3.7 software. d, e qRT-PCR (d) and IHC (e) analyses of TRAIL in OA articular cartilage from DMM surgery mice compared to sham-operated mice. Scale bar = 50 μm. f, g mRNA levels (f) and immunostaining (g) for OPG in articular cartilage tissue from sham surgery or DMM surgery mice. Scale bar = 50 μm. h, i Apoptotic articular chondrocytes were detected and quantified by TUNEL assay. Scale bar = 25 μm. Error bars represent mean ± S.E.M. of n = 10 mice (d–g, i). Two-way ANOVA was performed followed by Sidak’s Multiple Comparison’s test, with p values indicated in figure. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32859940), licensed under a CC-BY license. Not internally tested by R&D Systems.Applications for Mouse TRAIL/TNFSF10 Antibody
Immunohistochemistry
Sample: Perfusion fixed frozen sections of mouse thymus
Formulation, Preparation, and Storage
Purification
Reconstitution
Formulation
Shipping
Stability & Storage
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: TRAIL/TNFSF10
TNF-related apoptosis-inducing ligand (TRAIL), also called apoptosis 2 ligand (Apo2L) for its similarity in sequence, structure, and function to Fas Ligand/Apo1L, is a 33-35 kDa type II transmembrane glycoprotein of the tumor necrosis factor superfamily, designated TNFSF10 (1-3). Mouse TRAIL cDNA encodes a 17 amino acid (aa) N-terminal intracellular domain, a 20 aa transmembrane domain and a 253 aa extracellular domain. Like most TNF family members, TRAIL is bioactive as a homotrimer (1). Unlike other TNF family members, a zinc ion complexed by human Cys 230 (mouse Cys 240) of each of the three monomers is critical for structural stability (4, 5). Either transmembrane or cysteine protease-released soluble sTRAIL induce apoptosis of many transformed cell lines, but rarely of normal cells (3, 6). Accordingly, TRAIL is suggested to have a role in tumor surveillance (1). Mice with genetically disrupted TRAIL have defective thymocyte apoptosis, creating faulty negative selection and some increased susceptibility to induced autoimmune diseases (7). In humans, TRAIL controls apoptosis of erythrocyte precursors and sTRAIL is inversely correlated with hemoglobin (1, 8). TRAIL transcripts are constitutively expressed in a variety of human (and presumably mouse) tissues and mononuclear cells (2, 3). Only one of two receptors that transduce apoptotic signals in humans is found in the mouse (TRAIL R2/DR5 but not TRAIL R1/DR4) (1). Mice express TRAIL receptors DcTRAIL R1/TNFRSF23, and DcTRAIL R2/TNFRSF22. These receptors lack death domains but differ in structure from human regulatory receptors TRAIL R3 and TRAIL R4 (9). Osteoprotegerin has been identified in humans as a TRAIL receptor, but binding in mouse has not yet been demonstrated (1, 10). Mouse TRAIL shows 85% aa identity with rat TRAIL and 70% aa identity with human, bovine, and porcine TRAIL within the TNF homology domain (aa 118-291).
References
- Zauli, G. and P. Secchiero (2006) Cytokine Growth Factor Rev. 17:245.
- Wiley, S.R. et al. (1995) Immunity 3:673.
- Pitti, R.M. et al. (1996) J. Biol. Chem. 271:12687.
- Bodmer, J.L. et al. (2000) J. Biol. Chem. 275:20632.
- Hymowitz, S.G. et al. (2000) Biochemistry 39:633.
- Sedger, L.M. et al. (2002) Eur. J. Immunol. 32:2246.
- Lamhamedi-Cherradi, S.E. et al. (2003) Nat. Immunol. 4:255.
- Choi, J.W. (2005) Ann. Hematol. 84:728.
- Schneider, P. et al. (2003) J. Biol. Chem. 278:5444.
- Emery, J. et al. (1998) J. Biol. Chem. 273:14363.
Long Name
Alternate Names
Gene Symbol
UniProt
Additional TRAIL/TNFSF10 Products
Product Documents for Mouse TRAIL/TNFSF10 Antibody
Product Specific Notices for Mouse TRAIL/TNFSF10 Antibody
For research use only