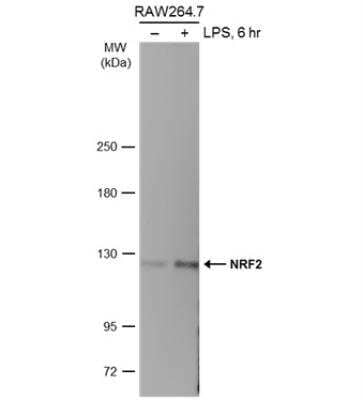
Western Blot Analysis of Nrf2 in Treated and Untreated RAW264.7 Whole Cell Extracts
Untreated (-) and treated (+) RAW264.7 whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membrane was blotted with NRF2 antibody [N2C2], Internal diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody (NBP2-19301) was used to detect the primary antibody.
Western Blotting of Nrf2 in Treated and Untreated Rat-2 Whole Cell Extracts
Untreated (-) and treated (+) Rat-2 whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membrane was blotted with NRF2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody (NBP2-19301) was used to detect the primary antibody.
Western Blotting of Nrf2 in Treated and Untreated Neuro2A Whole Cell Extracts
Untreated (-) and treated (+) Neuro2A whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membrane was blotted with NRF2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody (NBP2-19301) was used to detect the primary antibody.
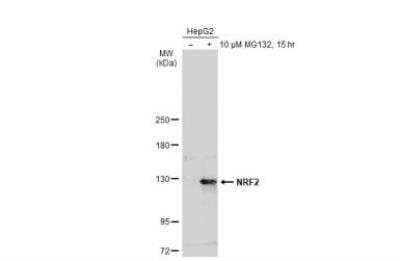
Western Blotting of Nrf2 in Treated and Untreated HepG2 hole Cell Extracts
Untreated (-) and treated (+) HepG2 whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membrane was blotted with NRF2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody (NBP2-19301) was used to detect the primary antibody.
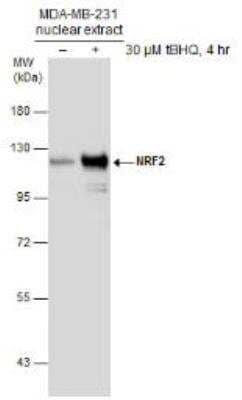
Western Blot Analysis of Nrf2 in Treated and Untreated MDA-MB-231 Nuclear Extracts
Untreated (-) and treated (+) MDA-MB-231 nuclear extracts (30 ug) were separated by 7.5% SDS-PAGE, and the membrane was blotted with NRF2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:1000.
Western Blot Analysis of Nrf2 in Treated and Untreated HepG2 Whole Cell Extracts
Untreated (-) and treated (+) HepG2 whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membranes were blotted with NRF2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:500 and competitor's antibody diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody (NBP2-19301) was used to detect the primary antibody.
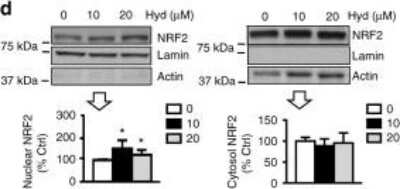
Detection of Nrf2 in Treated SH-SY5Y Cells by Western Blot
Hydralazine enhances NRF2 signaling in SH-SY5Y cells d NRF2 translocates to the nucleus with hydralazine treatment. Treated cells were subjected to cell fractionation and western blot analysis. *p < 0.05 and **p < 0.01, two-tailed Student's t test, n = 3, mean +/- SD. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/s41467-017-02394-3), licensed under a CC-BY license.
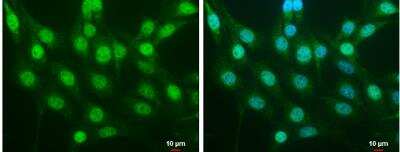
Immunocytochemistry/Immunofluorescence Analysis of Nrf2 in NIH/3T3 Cells
NIH/3T3 cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: NRF2 protein stained by NRF2 antibody [N2C2], Internal diluted at 1:500. Blue: Hoechst 33342 staining. Scale bar = 10 um.
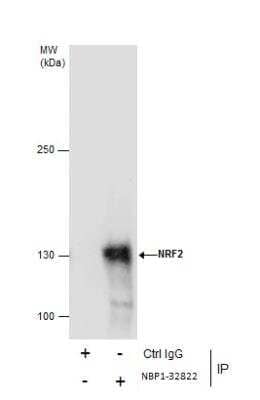
Immunoprecipitation of Nrf2 from HepG2 Whole Cell Extracts Using Nrf2 Antibody
Immunoprecipitation of NRF2 protein from HepG2 whole cell extracts using 5 ug of NRF2 antibody [N2C2], Internal Western blot analysis was performed using NRF2 antibody [N2C2], Internal.. EasyBlot anti-Rabbit IgG was used as a secondary reagent.
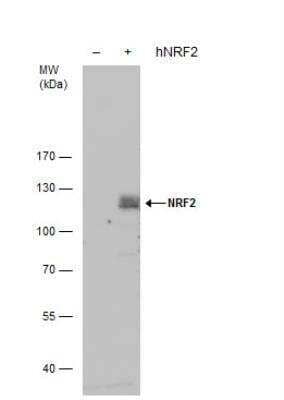
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - Blueberry polyphenol extract (BPE) increases the expression of NRF2 & HO-1 while reducing NF-kappa B p65 phosphorylation in angiotensin (Ang) II-treated human aortic endothelial cells (HAECs). HAECs were treated with 200 µg/mL of BPE for 1 h then treated with 200 nM of Ang II for 12 h. Protein expression of NRF2 (A,B), HO-1 (C,D), & NF-kappa B p65 (E,F) were determined by Western blot. Quantification was performed using Image Lab (Bio-Rad Laboratories, Inc.). Data are expressed as mean ± SD from nine (HO-1), & three (NRF2 & NF-kappa B) independent experiments. Values that do not share the same letter are significantly different from each other (p ≤ 0.05). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/35453301), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - NRF2 antibody validation. (A) NRF2 expression was silenced in MCF7 cells by transiently transfecting NRF2-specific siRNAs or a negative control siRNA for 48 h, then NRF2 protein expression was determined using two different antibodies (Abcam: ab31163; NOVUS: NBP1-32822). (B–D) 4T1 cells were treated with NRF2 activators, RA839 or tBHQ, or MG-132, a proteasome inhibitor, in the concentrations indicated for 48 h, then NRF2 protein expression was determined by western blotting using two different antibodies (Abcam: ab31163; NOVUS: NBP1-32822). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31461945), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - Maternal exercise during pregnancy on mitochondrial biogenesis in the fetal hearts. (A) Levels of relative mRNA expression measured by qRT‐PCR. n = 9–12/group. Maternal exercise during pregnancy did not alter levels of mRNA in Ppargc1a & Tfam, while it significantly upregulated the levels of mRNA in Nrf1 & Nrf2. (B–D) Densitometric analyses of protein expression levels relative to the sedentary group with representative images of western blots were shown. No significant differences in PGC‐1 alpha, NRF1, & NRF2 (P > 0.05). n = 5–6/group. * P < 0.05, significantly different from the sedentary group. Black bar: fetal hearts from sedentary dams; gray bar: fetal hearts from exercised dams. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/28292876), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 is required for UroA/UAS03 mediated upregulation of tight junction proteins. a Nrf2 levels were determined by immunoblots in HT29 cells treated with vehicle/UroA/UAS03 (50 μM) for 24 h. b Nrf2 expression in cytosolic & nuclear fractions of HT29 cells treated with Veh/UroA/UAS03 (50 μM) for 6 h. c Immunofluorescence confocal images of HT29 cells treated with vehicle/UroA/UAS03 (50 μM) for 6 h. The cells were stained with anti-Nrf2 antibody & DAPI. Relative green fluorescence (n = ~20 cells) intensity was measured. Scale bars indicate 25 μm. d Expression of Cldn4 & NQO1 in colon explants from WT, Nrf2−/−, & AhR−/− mice treated with vehicle/UroA/UAS03 (50 μM) for 24 h. Immunoblots were quantified using Image J software. e mRNA levels of Cldn4, Nrf2, & HO1 from colon explant cultures was measured by real-time PCR using SyBr green method. f C57BL/6, Nrf2−/−, & AhR−/− mice (n = 3) treated orally daily with veh or UroA/UAS03 (20 mg/kg) for 1 week. Cldn4 & NQO1 protein levels in colons were measured by immunoblots & quantified by Image J software. All in vitro studies were performed in triplicates. The immunoblots of colon explants & colon tissues were quantified from at least 6 independent runs. The levels of proteins were normalized to beta-actin & Wild type vehicle treatment was set to 1 & calculated the fold changes. Statistics performed using unpaired t-test using Graphpad Prism software. Error bars, ±SEM; *p < 0.05; **p < 0.01; ***p < 0.001. Source Data are provided as a Source Data File Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30626868), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - Modulation of Nrf2 & Keap1 mRNA & protein levels by compounds 1–6, curcumin (CURC), & dimethyl fumarate (DMF). (A–B) RNA from total cellular extracts of SH-SY5Y cells treated for 24 hours with 5 μM compounds or 20 μM DMF were analyzed for Nrf2 (A) & Keap1 (B) mRNA expression by RT-qPCR. GAPDH was used as housekeeping gene. Results are shown as mean ± SEM; no statistically significant data with Dunnett’s multiple comparison test (A, n = 3, F ratio = 1.249; B, n = 3, F ratio = 1.671). (C–D) Cellular extracts of SH-SY5Y cells treated for 24 hours with compounds at 5 μM or 20 μM DMF were analyzed for Nrf2 (C) & Keap1 (D) protein levels by Western blot. Anti-tubulin was used as protein loading control. Results are shown as ratio (% of CTR) ± SEM; **p < 0.01, versus CTR; Dunnett’s multiple comparison test (C, n ≥ 5, F ratio = 3.981; D, n = 3, F ratio = 0.4049). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32047434), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - CT activated AMPK/SIRT1 signaling. (A,B) HepG2 cells were treated with 2.5 μM CT for indicated times. Western blot analysis of phosphorylated AMPK, ACC, SIRT1, & Nrf2. (C) C57BL/6 mice were pair-fed either control or ethanol-containing diet with or without CT (20 or 40 mg/kg) for four weeks. Western blot analysis of phosphorylated AMPK, SIRT1. CYP2E1, & Nrf2. (D) HepG2 cells were incubated with 50 mM ethanol & treated with CT (2.5 or 5 μM) for 24 h. Western blot analysis of phosphorylated AMPK, SIRT1, CYP2E1, & Nrf2. (E) AML-12 cells were incubated with 50 mM ethanol & treated with CT (2.5 or 5 μM) for 24 h. Western blot analysis of phosphorylated AMPK & SIRT1. The images are representative (F) HepG2 cells were pretreated with CT (2.5 μM) for 3 h or with compound C (comp C) (10 μM) for 6 h, followed by ethanol (100 μM) treatment. Measurement of intracellular TG levels. Data are shown as mean ± SD of three independent experiments. #p < 0.05 vs. untreated control, ** p < 0.01 vs. ethanol-treated group. §§p < 0.01 vs. ethanol & CT-treated group. Densitometric analysis of western blots are given in Supplementary Figures S2 & S3A–G. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31906014), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - MPE & MSE exert anti-oxidant effects in 3T3-L1 adipocytes. 3T3-L1 cells were treated with pro-differentiative agents for 8 days in the presence or absence of 100 µg/mL MPE or MSE, as reported in Methods. (A) Intracellular ROS were detected using the redox-sensitive fluorochrome H2-DCFDA. After differentiation, the medium was replaced with 10 µM H2DCFDA solution & the incubation was protracted for 30 min at 37 °C. The oxidation of the fluorochrome generates green fluorescence, which was visualized by a Leica microscope equipped with a DC300F camera using a FITC filter. Representative micrographs of fluorescence microscopy were taken at 200× magnification. (B). Western blotting analysis of Nrf2, MnSOD & HO-1 in 3T3-L1 cells differentiated without or with 100 µg/mL MPE or MSE. Equal loading of proteins was verified by immunoblotting for beta-actin & showed values were assigned in relation to undifferentiated cells (Undif.). The bar graphs represent the mean of three independent experiments ± SD. * p < 0.05, ** p < 0.01 with respect to the undifferentiated 3T3-L1 cells, # p < 0.05, ## p < 0.01 & ### p < 0.001 with respect to the differentiated untreated 3T3-L1 cells (Dif.). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/35204243), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - NRF2 modulates epirubicin resistance in breast cancer cells. (A) MCF-7 & MCF-7 EpiR cells were treated for 24 h with epirubicin at 1 µM. MCF-7 & MCF-7 EpiR cells were stained with CellROX Deep Red reagent & analyzed for ROS levels by flow cytometry. Data were analyzed by FlowJo software. The mean fluorescence values were presented as relative ROS level compared to untreated cells (0 µM). Data presented as mean ± SD. Student’s t-test was used to compare the means: ** p < 0.01; *** p < 0.001; n.s. nonsignificant. (N = 4) (B) Knockdown of NRF2 was achieved by transfecting 4 specific siRNA against NRF2 (siNRF2, 150pmol) to MCF-7 EpiR cells in a 6-well plate. Non-targeting siRNAs were used as control (NSC). At 24 h post-transfection, these cells were seeded in 6-well plates & treated with increasing doses of epirubicin for 14 days. Their sensitivity to epirubicin was assessed by clonogenic assay. Their clonogenicity in response to epirubicin was analyzed by two-way ANOVA & found to be significantly different (** p < 0.01) from one another. (N = 3) (C) Expression of NRF2 in MCF-7 cells & MCF-7 EpiR cells was detected by Western blot. beta-Tubulin served as the loading control. (N = 3). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32110852), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: Nrf2 Antibody [NBP1-32822] -
Immunocytochemistry/ Immunofluorescence: Nrf2 Antibody [NBP1-32822] - Nrf2 antibody [N2C2], Internal detects Nrf2 protein at nucleus by immunofluorescent analysis.Sample: Neuro2A cells were fixed in 4% paraformaldehyde at RT for 15 min.Green: Nrf2 stained by Nrf2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:1000.
Immunohistochemistry-Paraffin: Nrf2 Antibody [NBP1-32822] -
Immunohistochemistry-Paraffin: Nrf2 Antibody [NBP1-32822] - Nrf2 antibody [N2C2], Internal detects Nrf2 protein at cytoplasm and nucleus by immunohistochemical analysis.Sample: Paraffin-embedded human breast carcinoma.Nrf2 stained by Nrf2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:500.Antigen Retrieval: Citrate buffer, pH 6.0, 15 min
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - Untreated (-) and treated (+) Rat2 whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membrane was blotted with Nrf2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:2000. The HRP-conjugated anti-rabbit IgG antibody was used to detect the primary antibody, and the signal was developed with Trident ECL plus-Enhanced.
Western Blot: Nrf2 Antibody [NBP1-32822] -
Western Blot: Nrf2 Antibody [NBP1-32822] - Untreated (-) and treated (+) HepG2 whole cell extracts (30 ug) were separated by 5% SDS-PAGE, and the membrane was blotted with Nrf2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:2000. The HRP-conjugated anti-rabbit IgG antibody was used to detect the primary antibody.
Immunocytochemistry/ Immunofluorescence: Nrf2 Antibody [NBP1-32822] -
Immunocytochemistry/ Immunofluorescence: Nrf2 Antibody [NBP1-32822] - Nrf2 antibody [N2C2], Internal detects Nrf2 protein at nucleus by immunofluorescent analysis.Sample: HeLa cells were fixed in 4% paraformaldehyde at RT for 15 min.Green: Nrf2 stained by Nrf2 antibody [N2C2], Internal (NBP1-32822) diluted at 1:1000.Red: phalloidin, a cytoskeleton marker, diluted at 1:200.Scale bar= 10 um.




![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202415293339.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-31020241534347.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202415175278.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202416163781.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202416163785.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202416165635.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202416171427.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-310202416165611.jpg)
![Immunocytochemistry/ Immunofluorescence: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-91020242021994.jpg)
![Immunohistochemistry-Paraffin: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-910202420115018.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-910202419593583.jpg)
![Western Blot: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-91020242063329.jpg)
![Immunocytochemistry/ Immunofluorescence: Nrf2 Antibody [NBP1-32822] - Nrf2 Antibody](https://resources.bio-techne.com/images/products/nbp1-32822_rabbit-polyclonal-nrf2-antibody-9102024202465.jpg)