Western Blot Detection of PINK1 in Multiple Cell Lines
Western blot image of PINK1 antibody (BC100-494) in multiple cells lines. Human HeLa (lane 1), Mouse NIH-3T3 (lane 2), L929 (lane 3) and Rat PC12 (lane 4) whole cell protein were separated by SDS-PAGE on a 7.5% polyacrylamide gel. Protein was transferred to PVDF membrane and probed with 2 ug/mL BC100-494 in 1% BSA and detected with an HRP-conjugated anti-rabbit secondary antibody using chemiluminescence. Observed molecular weight ~55 kDa (arrowhead).
Western Blot Analysis of PINK1 in Mouse Liver and Hypatocytes
Analysis of PINK1 in mouse liver and hypatocytes using PINK1 antibody. Observed molecular weight ~55 kDa. WB image submitted by a verified customer review.
Detection of PINK1 in Western Blot Using PINK1 Antibody
Pathogenic mutants of Parkin are subjected to Ser65 phosphorylation. Phos-tag Western blotting for Parkin and Western blotting for PINK1 were performed using Parkin WT and a series of pathogenic mutants. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/srep01002) licensed under a CC-BY license.
Western Blot Analysis of PINK1 in Transfected SH-SY5Y Cells
Alteration of mitochondria and PD associated proteins in SH-SY5Y cells with telomere removal by CRISPR-Cas9. Representative Western blot of PGC-1alpha, NRF1, and PINK1 in SH-SY5Y cells transfected with either gTel or gCont (72 h). Beta-actin served as a loading control. Image collected and cropped by CiteAb from the following publication (https://www.mdpi.com/1422-0067/18/10/2093), licensed under a CC-BY license.
Western Blot Detection of PINK1 in Neuro2A Whole Cell Lysate
35 ug Neuro2A whole cell lysate separated on 8% PAGE and stained for PINK1 (1:1000 in 5% milk powder in TBST; secondary antibody AP at 1:5000). WB image submitted by a verified customer review.
Immunohistochemical Staining of PINK1 in Paraffin Embedded Rabbit Heart Tissue
Rabbit heart tissue. IHC-P image submitted by a verified customer review.
Knockdown Validation of PINK1 Antibody in Treated and Untreated siRNA Transfected HEK293 Cells
HEK293 cells were co-transfected with PINK1 siRNA (#1 or #2) or scrambled siRNA (scrambled) and untagged wild-type (WT) or Ser65Ala (S65A) mutant Parkin as indicated using TransFectin reagent (Bio-Rad). 48 hrs post-transfection, cells were treated with/without 10 uM CCCP for 3 h. 0.25 mg of 1% Triton whole-cell lysate were subjected to immunoprecipitation with GST-Parkin antibody (S966C) covalently coupled to protein G Sepharose and then immunoblotted with anti-phospho-Ser65 antibody in the presence of dephosphorylated peptide. 5% of the IP was immunoblotted with total anti-Parkin antibody. 0.25 mg of whole-cell lysates were immunoprecipitated with anti-PINK1 antibody (S085D) and immunoblotted with anti-PINK1 antibody. Representative of three independent experiments. Image collected and cropped by CiteAb from the following publication (https://rsob.royalsocietypublishing.org/cgi/doi/10.1098/rsob.120080) licensed under a CC-BY license.
Knockout Validation of PINK1 Antibody in Treated/Untreated WT and PINK1 KO Mouse Platelets
Characterisation of mitochondrial content and function in PINK1-/- platelets. Cropped immunoblots showing PINK1 expression in 10 uM CCCP (6 hours) treated WT, but not KO platelets by IP (upper panel) with loading control of IP inputs by blotting for PINK1 (lower panel). Uncropped blots for PINK1 are provided in Supplementary Fig. S1b. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-018-32716-4), licensed under a CC-BY license.
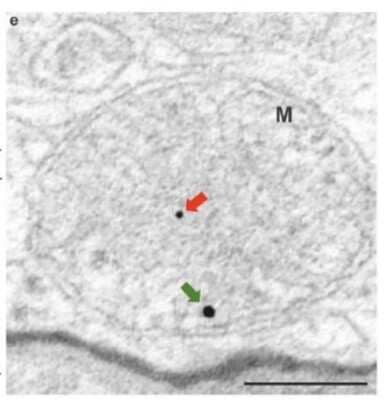
Electron Microscopy Detection of PINK1 in Mouse RPE Cells
Representative TEM picture. The red and green arrows indicate PINK and PARKIN immunogold particles in the dKO RPE samples, respectively (e). ML, melanosome. Scale = 0.2 um. Mitochondria (M). Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32183173) licensed under a CC-BY license.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Protein content and oxidation. Gastrocnemius muscle was analyzed by immunoblotting (A) for oxidatively modified protein, STIM1, PARKIN, PINK1, PGC‐1 alpha, VDAC1, and 4HNE (B). Data are means ± SEM. *Significantly different from Tp53inp1+/+.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PPM1H does not dephosphorylate Ser111 of Rab8A or key phosphorylation sites of AMPK & Akt signaling pathways.(A) HEK293 cells were transiently transfected with constructs expressing the indicated components. 24 hr post-transfection, cells were treated ±10 µM CCCP (Carbonyl cyanide m-chlorophenyl hydrazine) for 3 hr to induce activation of the PINK1 kinase & trigger Rab8A phosphorylation at Ser111 (Lai et al., 2015). (B) As in (A) except cells were immunoblotted with the indicated antibodies that recognize key phosphorylation sites of AMPK & Akt signaling pathways. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31663853), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 & Parkin are phosphorylated after a decrease in deltaΨm in mouse primary neurons. Neurons were infected with lentivirus encoding PINK1-Flag (A), wild-type HA-Parkin (B) or HA-Parkin with either the S65A or S65E mutation (C). Cells were treated with the mitochondrial uncoupler CCCP (30 μm) for 1–3 h & subjected to SDS-PAGE in the absence or presence of 50 μm phos-tag. Note that mobility does not reflect the molecular weight of proteins in phos-tag PAGE (Kinoshita et al. 15), & thus, molecular weight markers are not shown in the bottom gels. The red & black asterisks in (C) indicate phosphorylation of Parkin at Ser65 & an additional minor phosphorylation site, respectively. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23751051), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 translation is critical for the accumulation of PINK1 upon mitochondrial damage. aPINK1 mRNA levels were not altered by mitochondrial damage. HeLa cells were incubated with 10 μM CCCP or combination of 10 μM oligomycin & 4 μM antimycin for 2 h & then, PINK1 mRNA levels were measured by qRT-PCR. PINK1 mRNA level was normalized to the corresponding GAPDH levels & quantified as a percentage of control (n = 8, t-test). b, c Translation is involved in PINK1 accumulation upon CCCP treatment. HeLa cells were first pre-incubated with 100 μM cycloheximide (CHX) for 2 h & then washed. Cells were further incubated with 10 μM CCCP with or without CHX for 2 h (n = 4, two-way ANOVA). Values are mean ± SEM (n.s. = non-significant, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Protein content & oxidation. Gastrocnemius muscle was analyzed by immunoblotting (A) for oxidatively modified protein, STIM1, PARKIN, PINK1, PGC‐1 alpha, VDAC1, & 4HNE (B). Data are means ± SEM. *Significantly different from Tp53inp1+/+. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31124296), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - TP53 knockout triggers a pro-mitophagic response.PINK1 (a,b, N = 8), LC3-2/LC3-1 (a,c, N = 8), p62 (a,d, N = 12), TIM23 (a,e, N = 10), TOM20 (a,f, N = 9), HSP60 (a,g, N = 16), optineurin (a,h, N = 9) & NDP52 (a,i, N = 8) protein levels were analyzed in control (p53+/+) or TP53-deficient (p53−/−) HAP1 cells as described in the Methods. Bars represent the means ± SEM of 3-4 independent experiments performed in triplicate & are expressed as percent of control (p53+/+) cells. Actin expression is provided as a representative gel loading control in a. Statistical analyses were performed with GraphPad Prism software by using unpaired Student’s t-test. Significant differences are: ** p < 0.01, ***p < 0.001 & ****p < 0.0001. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29352272), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Mito-LND modulates energetic & autophagy signaling proteins in vitro & in vivo. a Mito-LND (2 µM) modulates AMPK & AKT/mTOR signaling cascades in H2030 & H2030BrM3 lung cancer cell lines over time. b Mito-LND (2 µM) modulates autophagy & specific mitophagy-linked proteins in H2030 & H2030BrM3 lung cancer cells. c Mito-LND (2 µM) modulates autophagy & specific mitophagy-linked proteins in A549 & NCI-H460 lung cancer cells. d Mito-LND induces autophagy in lung tumor tissues in mouse orthotopic model of lung cancer. e Induction of autophagy by Mito-LND in brain tumor tissues in the mouse brain lung metastasis model. The relative band intensities as determined by densitometry are indicated above each blot Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31101821), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Impairment of mitophagy upon Antimycin A & Oligomycin exposure.a Cells were treated with Antimycin A (1 μM) & Oligomycin (1 μM) for 1, 3, & 6 & mitochondrion/cytosol fractionation was performed to assay the accumulation of LC3-II, p62 & PINK1 within the mitochondrial fraction. VDAC1 was used as a loading control for mitochondrial fractions. b, c Cells were treated with Antimycin A (1 μM) & Oligomycin (1 μM) for 1, 3, 6, & 24 h & total fibroblasts extracts were analyzed for PINK1, p62, COXIV & VDAC1. Each quantitative data was normalized with ACTIN (n ≥ 3). Statistical analysis was performed using two-way ANOVA with Tukey’s multiple comparisons test. (*p < 0.05; **p < 0.01; ***p < 0.001). Immunoblots reported are from one representative experiment Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31332166), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - miR-27a/b prevent pUbS65 accumulation upon mitochondrial damage. a, b Overexpression of PINK1 increased pUbS65 accumulation in HeLa cells only when incubated with CCCP. 48 h post-transfection, cells were incubated with 10 μM CCCP for 2 h & pUbS65 levels were determined by Western blot (n = 3, two-way ANOVA). c, d Overexpression of miR-27a/b inhibited pUbS65 accumulation by CCCP in HeLa cells (n = 4, two-way ANOVA). e, f Inhibition of endogenous miR-27a/b increased pUbS65 accumulation by CCCP in HeLa cells (n = 4, two-way ANOVA). PINK1 levels were normalized to corresponding GAPDH level & quantified as a percentage of control. Values are mean ± SEM (n.s. = non-significant, **p < 0.01, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - The HIF-1 alpha-BNIP3 pathway was activated during hypoxia. (a) Analysis of mitophagy-related protein expression in Neuro-2a cells by immunoblotting. Neuro-2a cells were exposed to hypoxia and/or high glucose for 48 h. Total cellular extracts were analysed by immunoblotting with antibodies against HIF-1 alpha, BNIP3, PINK1 & TOMM20. (b) The ImageJ densitometric analysis is shown. “C” represented control conditions (25 mM glucose), “G” represented high glucose conditions (75 mM glucose). The results are expressed as the mean ± SEM from three independent experiments. Two-way ANOVA, **p < 0.01, *p < 0.05, Hypoxia vs Normoxia. Full-length blots are presented in Supplementary Fig. S2. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-018-20162-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - miR-27a/b inhibit PINK1 accumulation upon mitochondrial damage. a, b Overexpression of miR-27a/b inhibited PINK1 accumulation upon mitochondrial damage. 48 h post-transfection, HeLa cells were treated with 10 μM CCCP or combination of 10 μM oligomycin & 4 μM antimycin as indicated for 2 h (n = 4, two-way ANOVA). c, d Inhibition of endogenous miR-27a/b increased PINK1 accumulation upon CCCP treatment. (n = 4, two-way ANOVA). PINK1 levels were normalized to corresponding GAPDH level & quantified as a percentage of control. Values are mean ± SEM (n.s. = non-significant, *p < 0.05, **p < 0.01, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: PINK1 Antibody - BSA Free [BC100-494] -
Immunocytochemistry/ Immunofluorescence: PINK1 Antibody - BSA Free [BC100-494] - Confocal microscopy analysis of the mitophagy initiation in the RPE cells by staining PINK1 & PARKIN. One-year-old WT & dKO mice focusing on the RPE cells in the vicinity of the optic nerve (a,e). PINK1 (b, red) & PARKIN (c, green) were double-stained & the merged image (d) was used to count the colocalized puncta from WT. Similarly, in dKO PINK1 (f, red) & PARKIN (g, green) were double-stained, & the merged image (h) was used to count the colocalized puncta. In dKO, we observed a ~7% decrease in the total number of puncta; however, the number of colocalizations was increased by ~118% (i). Scale = 5 µm. *p = 0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/32183173), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Alteration of PARKIN leads to an impairment of mitophagy upon CCCP treatment.a 2 N & DS treated w/ CCCP (20 μM) for 6 h & mitochondrion/cytosol fractionation performed to assay the accumulation of LC3-II & p62 w/in mitochondrial fraction. VDAC1 & ACTIN used as a loading control for mitochondrial & cytosolic fractions, respectively. b, c Quantitative data of LC3-II & p62 in mitochondrial fraction normalized w/ VDAC1 (n = 3). d The cells treated w/ CCCP (20 μM) for 6 h & the whole-cell extracts from 2 N & DS analyzed by WB for PARKIN & PINK1. e Quantification of PARKIN normalized w/ ACTIN (n ≥ 3). d Quantification of PINK1 after 6 h of CCCP, each quantitative data normalized w/ ACTIN (n ≥ 3). g Representative confocal images of cells stained w/ Mitotracker green (mitochondrial marker) & Lysotracker red (lysosomal marker) upon CCCP treatment (20 μM for 6 h). White arrow heads point to co-localization w/in puncta from boxed region at high magnification shown on the right. Scale bar 10 μm. h The graph the % of Mitotracker green /Lysotracker red colocalization calculated by JACoP plugin of ImageJ. Minimum 50 cells per condition counted from three independent experiments. i 2 N & DS cells treated w/ CCCP (20 μM) for 6 h & assessed for SQSTM1 (p62), PARKIN & PINK1 mRNA by quantitative real-time PCR. mRNA levels normalized to ACTIN mRNA, used as internal control. Data display the fold-changes of SQSTM1, PARKIN & PINK1 mRNAs relative to control cells (n = 3). Statistical analysis performed using Student’s T-test or one-way ANOVA w/ Tukey’s multiple comparisons test. (*p < 0.05; **p < 0.01; ***p < 0.001; n.s. no significant difference). Immunoblots reported are from one representative experiment Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31332166), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: PINK1 Antibody - BSA Free [BC100-494] -
Immunocytochemistry/ Immunofluorescence: PINK1 Antibody - BSA Free [BC100-494] - Polyphyllin I triggers PINK1/PARK2-dependent mitophagyA. MDA-MB-231 cells treated w/ 8 μM PPI for different periods of time as indicated, & PARK2, P62, & ubiquitin (UB) levels in mitochondrial fractions determined by western blot. B. Cells cotransfected w/ GFP-UB & RFP-mito & treated w/ 8 μM PPI for 9 h, after which PARK2 (Alexa Fluor 405, pink) & P62 (Alexa Fluor 647, blue) immunostaining detected using confocal microscopy. C. Cells cotransfected w/ GFP-UB & RFP-LC3 treated w/ 8 μM PPI for 9 h, after which PARK2 (Alexa Fluor 405, pink) & TOMM20 (Alexa Fluor 647, blue) immunostaining detected using confocal microscopy. Scale bars: 10 μm. D-E. MDA-MB-231 cells treated w/ 8 μM PPI for different periods of time as indicated; whole-cell lysates then separated on 8% SDS-PAGE gels & analyzed by western blot using the anti-PINK1 antibody. Relative full-length (∼63 kDa) & cleaved (∼52 kDa) PINK1 levels quantified by densitometry & normalized to Tubulin. Results expressed as a percentage of control, which set at 100%. Data are presented as mean ± SD (*P< 0.01 vs. the control). F. Cells treated w/ 8 μM PPI for different periods of time as indicated, & whole-cell lysates then subjected to western blot analysis. G. Cells treated w/ 8 μM PPI for 9 h, after which mitochondrial fractions prepared & subjected to immunoprecipitation using anti-PINK1 antibody; associated PARK2 detected using immunoblotting. H. RFP-mito-expressing MDA-MB-231 cells treated w/ 8 μM PPI for 9 h, & PINK1 (Alexa Fluor 488, green) & PARK2 (Alexa Fluor 405, pink) immunostaining evaluated using confocal microscopy. Scale bars: 10 μm. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.14413), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 is required for Parkin translocation but interaction between Beclin1 & Parkin is PINK1 independent. (A) HEK293T cells were transfected with scrambled siRNA & siRNA targeting SESN2 followed by treatment with 10 µM CCCP for 3 h. Total cell lysates were co-immunoprecipitated using anti-PINK1 & anti-Parkin antibodies, to find the variation in interaction between endogenous PINK1 & Parkin. Interaction of PINK1 with Parkin is less prominent on downregulation of SESN2. (B) Effect of PINK1 on mitochondrial translocation of Parkin was assessed by transfecting cells with scrambled siRNA & PINK1 siRNA followed by treatment with 10 µM CCCP for 3 h. Cellular sub-fractionation was done & mitochondrial fraction was subjected to immunoblotting. TOMM20, mitochondrial marker. 1-DMSO, 2-CCCP. Note that PINK1 knockdown abolishes the presence of Parkin. (C) To check the mitochondrial stabilization of PINK1 in SESN2 knockdown cells, cells were treated with DMSO (vehicle) & CCCP for 3 h & western blot analysis was carried out in isolated mitochondrial fractions. VDAC1, mitochondrial marker & TUBB/ beta-Tubulin, cytosolic marker. Levels of SESN2 in PINK1 knockdown cells were analysed with western blots in whole cell lysates. 1-DMSO, 2-CCCP. Note: downregulation of SESN2 or PINK1 was unable to decline stability of each other. (D) Wild-type (WT) & PINK1 knockdown cells were treated with DMSO (vehicle) & CCCP for 3 h. Cytosolic fractions were isolated & subjected to co-IP assay using anti-Beclin1 & anti-Parkin antibodies. Immunoblots were converted to gray & white scale using Adobe Photoshop CS6, with linear adjustment of exposure or contrast. Images of immunoblots were cropped for clarity, expanded view of Fig. 6D is available in supplementary figures. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29330382), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - G-TPP but not 17-AAG induces Parkin translocation(A, B) HeLa cells stably expressing EGFP-Parkin were treated with 12 different concentrations of G-TPP or 17-AAG in a dose-response format for 2 or 6 h before adding (A) DMSO or (B) CCCP (final assay concentration = 10 µM) for an additional 2 h. Cells were fixed & analyzed for Parkin translocation (black line). Cell number was assessed by counting the number of Hoechst-positive nuclei in each well (gray line). Values were normalized to positive (2 h 10 µM CCCP) & negative (2 h DMSO) controls. In absence of CCCP, only G-TPP but not 17-AAG induced Parkin translocation. In combination with CCCP, increasing doses of 17-AAG led to inhibition of Parkin translocation. Very high concentrations of G-TPP also inhibited Parkin translocation & resulted in cell toxicity. (C) HeLa cells expressing untagged Parkin were treated with 1 µM 17-AAG, 10 µM G-TPP or DMSO as a control for 6 h before CCCP (10 µM) or medium containing DMSO was added for 4 h. Cells were harvested & western blots probed with antibodies against PINK1, pS65-Ub, the mitochondrial phosphatase PGAM5. Vinculin was used as a loading control. Compared to controls, 17-AAG pre-treated cells showed lower PINK1 levels that were accompanied by reduced pS65-Ub induction upon CCCP treatment, while G-TPP pre-treatment led to induction of pS65-Ub in the absence of CCCP, as expected. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.22287), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Knockdown of Pink1 inhibits mitophagy flux in CCCP-treated RPTC cells. (a) RPTC cells were subjected to: (1) control; (2) sIPC; (3) CCCP-R; (4) sIPC + CCCP-R. After treatment mitochondrial fractions were collected for immunoblot analysis of multiple mitophagy-related proteins including PINK1, PRKN, BNIP3L/NIX, FUNDC1 & DNM1L. HSPD1, a mitochondrial matrix protein, was used as a loading control. (b) RPTC cells were infected with retroviral Pink1 shRNA constructs (A–D) & a negative control (NC) construct. Upon puromycin selection, stable cells were collected for immunoblot analysis of PINK1. PPIB was used as a loading control. Based on the inhibitory effects, the stable cells (negative control, Pink1 shRNA A, Pink1 shRNA C) were transfected with COX8-EGFP-mCherry & then treated with: (1) control; (2) CCCP-R; (3) sIPC + CCCP-R. Cells were collected for fluorescence microscopy. (c) Representative images of mitolysosome formation. Scale bar: 10 μm. (d) Quantitative analysis of the number of mitolysosomes per cell. Data are expressed as mean ± SD. *, P < 0.05, significantly different from the control group; #, P < 0.05, significantly different from CCCP-R group; ^, P < 0.05, significantly different from the corresponding groups in negative control cells. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31066324), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - G-TPP triggers PINK1 activation in iNeurons(A) Control cells or PINK1 Q456X fibroblasts were converted to induced neurons (iNeurons). Cells were treated for the indicated times with 10 µM G-TPP & harvested. Western blots were prepared & probed with antibodies against PINK1, pS65-Ub & Parkin. Beta III tubulin served as a control for successful conversion to neuronal cells, GAPDH as a loading control. (B) iNeurons were treated with 15 µM G-TPP for 8 h & fixed. Cells were stained with antibodies against pS65-Ub (green) & the mitochondrial marker TOM20 (red) & the neuronal marker Beta III tubulin (cyan). Nuclei were stained with Hoechst 33342 (blue). Scale bars correspond to 10 µM. A magnified image of the boxed region & the fluorescence profile along the arrow are shown to the right. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.22287), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - miR-27a/b suppress human PINK1 expression. a Schematic diagram of the conserved target sites of miR-27a/b in the 3′UTR of human PINK1 mRNA. Seed sequences were indicated in red. Asterisks (*) indicate the conserved nucleotides. b Schematic diagram of the luciferase reporter plasmids. The reporter constructs contain the full-length 3′UTR of human PINK1 mRNA with wild-type (WT) or mutated (Mut) seed match sites downstream of Renilla luciferase gene. c Overexpression of miR-27a/b decreased luciferase activities in HeLa cells. Cells were transfected with miR-27a/b or negative control (Ctl) along with reporter constructs as indicated. 48 h post-transfection, luciferase activities were measured (n = 4, one-way ANOVA). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity. d-g Overexpression of miR-27a/b decreased PINK1 protein levels in HeLa (D-E) & M17 cells (F-G). Cells were transfected with miR-27a/b or negative control (Ctl). 48 h post-transfection, cells were harvested for Western blot (n = 4, t-test). PINK1 protein level was normalized to corresponding GAPDH level & quantified as a percentage of control. Data are shown as a percentage of control. Values are mean ± SEM (n.s. = non-significant, **p < 0.01, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Polyphyllin I triggers PINK1/PARK2-dependent mitophagyA. MDA-MB-231 cells were treated with 8 μM PPI for different periods of time as indicated, & PARK2, P62, & ubiquitin (UB) levels in mitochondrial fractions were determined by western blot. B. Cells were cotransfected with GFP-UB & RFP-mito & treated with 8 μM PPI for 9 h, after which PARK2 (Alexa Fluor 405, pink) & P62 (Alexa Fluor 647, blue) immunostaining was detected using confocal microscopy. C. Cells cotransfected with GFP-UB & RFP-LC3 were treated with 8 μM PPI for 9 h, after which PARK2 (Alexa Fluor 405, pink) & TOMM20 (Alexa Fluor 647, blue) immunostaining was detected using confocal microscopy. Scale bars: 10 μm. D-E. MDA-MB-231 cells were treated with 8 μM PPI for different periods of time as indicated; whole-cell lysates were then separated on 8% SDS-PAGE gels & analyzed by western blot using the anti-PINK1 antibody. Relative full-length (∼63 kDa) & cleaved (∼52 kDa) PINK1 levels were quantified by densitometry & normalized to Tubulin. The results were expressed as a percentage of control, which was set at 100%. Data are presented as mean ± SD (*P< 0.01 vs. the control). F. Cells were treated with 8 μM PPI for different periods of time as indicated, & whole-cell lysates were then subjected to western blot analysis. G. Cells were treated with 8 μM PPI for 9 h, after which mitochondrial fractions were prepared & subjected to immunoprecipitation using anti-PINK1 antibody; associated PARK2 was detected using immunoblotting. H. RFP-mito-expressing MDA-MB-231 cells were treated with 8 μM PPI for 9 h, & PINK1 (Alexa Fluor 488, green) & PARK2 (Alexa Fluor 405, pink) immunostaining were evaluated using confocal microscopy. Scale bars: 10 μm. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.14413), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Inhibition of endogenous miR-27a/b function increases human PINK1 expression. a Schematic diagram of sequence alignment of miR-27a/b & anti-miR-27a/b. Asterisks (*) indicate conserved nucleotides. b Inhibition of endogenous miR-27a/b function increased luciferase activities in HeLa cells. Cells were transfected with the reporter constructs containing the full-length 3′UTR of human PINK1 mRNA with wild-type (WT) or mutated (Mut) seed match sites downstream of Renilla luciferase gene. 48 h post-transfection, luciferase activities were measured (n = 4, t-test). c-f Inhibition of endogenous miR-27a/b increased PINK1 protein levels in HeLa (C-D) & M17 cells (E-F). Cells were transfected with 150 nM of anti-miR-27a/b or anti-control (anti-Ctl). 48 h post-transfection, cells were harvested for Western blot. PINK1 protein level was normalized to corresponding GAPDH level & quantified as a percentage of control (n = 4, t-test). Data are shown as a percentage of control. Values are mean ± SEM (n.s. = non-significant, **p < 0.01, ***p < 0.00) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 knockdown combined w/ polyphyllin I treatment blocks mitophagy & increases mitochondrial fission & apoptosisA. MDA-MB-231 cells stably expressing non-target shRNA (shCon) or PINK1 shRNA (shPINK1) lysed & analyzed by western blot. B-C. shCon & shPINK1 cells treated w/ or w/out 8 μM PPI for 6 h; whole-cell lysates & mitochondrial fractions then prepared & subjected to western blot analysis. D-E. shCon & shPINK1 cells cotransfected w/ RFP-mito & GFP-LC3, & then treated w/ 8 μM PPI for 6 h. LAMP1 (Alexa Fluor 647, blue) immunostaining then detected using confocal microscopy. Scale bars: 10 μm. The percentage of cells in which mitophagy occurred determined using 30 cells from each experiment; 3 independent experiments are included. The cells w/ more than five RFP-Mito, LC3, & LAMP1 colocalization puncta designated mitophagy-positive. Data are presented as mean ± SD (*P< 0.01 compared to shCon cells treated w/ PPI). F-G. shCon & shPINK1 cells transfected w/ RFP-mito, & then treated w/ 8 μM PPI for 6 h. Mitochondria observed using confocal microscopy. Scale bars: 10 μm. The average mitochondrial length quantified as previously described. Data are presented as mean ± SD (*P< 0.01 compared to shCon cells treated w/ PPI). H. shCon & shPINK1 cells treated w/ 8 μM PPI for 6 h, & DRP1 levels in mitochondrial fractions determined by immunoblotting. I. Cells treated w/ or w/out 8 μM PPI for 6 h, & apoptosis then measured by flow cytometry. Data are presented as mean ± SD (*P< 0.01 compared to shCon cells treatment w/ PPI). J-K. Whole-cell lysates, mitochondrial (Mito), & cytosolic (Cyto) fractions prepared & subjected to western blot analysis. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.14413), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Mitophagy execution via PINK1 accumulation, MFN2/TOM20 reduction & mitophagy signaling in MDA-MB-231 cells in vitro & SST2 tumors in vivo(A) Confocal images of GFP-LC3 expressing MDA-MB-231 cells preloaded with MitoTracker Red at 12 hours post MTA treatment. Scale bar is 10 μm. Yellow box indicates the area selected for obtaining z stacks. (B) Cropped immunoblot of the outer mitochondrial membrane proteins MFN2, TOM20 & VDAC in MDA-MB-231 cells treated with 1 μM of different MTAs at the indicated times. Bars represent the mean ± SD SEM. (n=3) (C) Representative cropped LC3 immunoblots of a mitochondrial extract (300 μg) after MFN2 immunoprecipitation from MDA-MB-231 cells exposed to 1 μM of different MTAs for 24 hours to confirm an endogenous protein complex interaction between the autophagosome & mitochondria. Bar represents the mean ± SEM. Two replicates (Rep.) are shown. (n=3) (D) Representative cropped PINK1 immunoblot from the mitochondrial fractions of cells treated with different MTAs for 12 hours. Bar represents the mean ± SEM. (n=4) (E) Representative cropped PINK1 immunoblot from tumor mitochondrial extracts using a rat SST-2 allograft model after DMSO or MitoQ treatment for 14 days. Bar represents the mean ± SD. (n=3) *P<0.05, & **P<0.01 indicate statistical significance. (F) Schematic model for the proposed mechanism for the mitophagy selective response induced by MTA in MDA-MB-231 cells as compared to MCF-12A cells (See Discussion). Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.23171), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Inactivation of ATF3 potentiates PINK1 transcription. (a) Representative immunoblot analysis of ATF3 & PINK1 in total cell lysates of A549 cells, transfected with GFP (transfection control) or ATF3. Cells overexpressing ATF3 for 48 hr show lower levels of PINK1 in whole cell lysates. A549 cells transfected with siRNA scramble control or ATF3 siRNA for a total of 48 hr & exposed to tunicamycin the last 24 hr (b–d). Less ATF3 mRNA after 24 hr TM treatment (b) & a recovery of the basal PINK1 transcript levels (c) were measured in knockdown ATF3 cells. (d) At 48 hr, protein levels of ATF3 also reflect these changes after TM treatment in the presence or absence of ATF3 silencing (see Figure S2F). Data represent mean ± SEM of four (a–c) & three (d) independent experiments. *p < .01, two‐way ANOVA with multiple comparison test Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29363258), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - miR-27a/b prevent Parkin translocation to mitochondria upon mitochondrial damage. a, b 48 h post-transfection, HeLa cells were incubated with 10 μM CCCP for 2 h. After isolating the mitochondria-enriched fractions, Parkin translocation & LC3-II were assessed by Western blot. Each protein level was normalized to corresponding mitochondrial loading control (SOD2) level & quantified as a percentage of control (n = 3, two-way ANOVA). Purity of mitochondrial fraction was assessed by monitoring p38 cytosolic & VDAC1 mitochondrial markers. PN; post-nuclear, C; cytoplasm, M; mitochondrial (A). c, d 48 h post-transfection, HeLa cells stably expressing GFP-Parkin were visualized by GFP, a mitochondrial marker (TOM20) antibody, & a nuclear dye (Hoechst) as indicated. Scale bars correspond to 10 μm. Parkin translocation was quantified as the ratio of cytoplasmic to nuclear GFP signal & the resulting ratios were normalized to control. Data were collected from 4 independent replicates (n > 1000 cells) & are shown as a percentage of control. Values are mean ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Inhibition of endogenous miR-27a/b function increases human PINK1 expression. a Schematic diagram of sequence alignment of miR-27a/b & anti-miR-27a/b. Asterisks (*) indicate conserved nucleotides. b Inhibition of endogenous miR-27a/b function increased luciferase activities in HeLa cells. Cells were transfected with the reporter constructs containing the full-length 3′UTR of human PINK1 mRNA with wild-type (WT) or mutated (Mut) seed match sites downstream of Renilla luciferase gene. 48 h post-transfection, luciferase activities were measured (n = 4, t-test). c-f Inhibition of endogenous miR-27a/b increased PINK1 protein levels in HeLa (C-D) & M17 cells (E-F). Cells were transfected with 150 nM of anti-miR-27a/b or anti-control (anti-Ctl). 48 h post-transfection, cells were harvested for Western blot. PINK1 protein level was normalized to corresponding GAPDH level & quantified as a percentage of control (n = 4, t-test). Data are shown as a percentage of control. Values are mean ± SEM (n.s. = non-significant, **p < 0.01, ***p < 0.00) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Regulation of PINK1 by miRNA. a Schematic diagram of AGO-specific RNA immunoprecipitation assay. b, cPINK1 mRNAs are associated with miRNA/RISC complex. HeLa cells were transfected with 50 nM of AGO2 siRNA or negative control. 48 h post-transfection, AGO2-bound mRNAs were pulled down from cell lysates with 2A8 anti-AGO2 antibody or non-specific IgG antibody. PINK1 mRNA levels were then measured by qRT-PCR. siRNA against AGO2 served as a quality control for AGO-specific RNA immunoprecipitation assay. PINK1 mRNA levels were quantified as a percentage of group 1 (n = 6, two-way ANOVA). d, e Knock-down of DICER1 increased PINK1 protein levels. HeLa cells were transfected with 50 nM DICER1 siRNA (DICER1 KD) or negative control (Ctl). 48 h post-transfection, cells were harvested for Western blot. The arrowhead indicates a full-length PINK1 band & the asterisk indicates a non-specific band. Each protein level was normalized to corresponding GAPDH level & quantified as a percentage of control (n = 4, t-test). Values are mean ± SEM (n.s. = non-significant, **p < 0.01, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - The expression levels of microtubule protein light chain 3 (LC3B), PTEN inducing putative kinase 1 (PINK1), Parkin, mitofusin 1 (Mfn1), toll-like receptor (TLR) 9, cytochrome c oxidase 4 (COX4), myeloid differentiation factor 88 (MyD88), & nuclear factor (NF)-kappa B in lung tissues from animals with spontaneous breathing (CON) or mechanical ventilation at high tidal volume (HTV) with saline, antimycin A (AmA) or cyclosporine A (CsA). (A) Levels of PINK1, Parkin, & Mfn1 mRNA. (B) Levels of LC3B, PINK1, Parkin, & Mfn1 protein by Western blot. (C) Relative expression of LC3B-II/LC3B-I & PINK1 protein. (D) Relative expression of Parkin & Mfn1 protein. (E) Levels of TLR9 & COX4 mRNA. (F) Levels of MyD88 & nuclear factor-kappa B (NF-kappa B) mRNA. (G) Levels of TLR9, COX4, MyD88, & NF-kappa B protein by Western blot. (H) Relative expression of TLR9 & COX4 protein. (I) Relative expression of MyD88 & NF-kappa B protein. Fold expression for target genes was normalized to that measured for the beta-actin gene. Both of these experiments were in triplicate. aP < 0.05 vs. CON group;bP < 0.05 vs. HTV group; & cP < 0.05 vs. AmA group. Image collected & cropped by CiteAb from the following publication (https://www.frontiersin.org/article/10.3389/fimmu.2018.01477/full), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Mitophagy contributes to removal of excess mitochondrial fragments in preeclampsia.(a, left panel) Representative TEM image of a cytotrophoblast from PTC & PE placenta collected at 29 weeks. Mitochondrial proximity to ER (MAM) is indicated by white arrows, mitochondrial fission events are depicted by white stars (scale bar: 500 nm; n = 8 separate PE placentae). (a, right panel) percentage of mitochondrial tethering to the ER in in PE vs. PTC (PE placentae, n = 8; PTC placentae, n = 7; unpaired Student’s t-test *P < 0.05). b Representative western blots for MFN2 & calreticulin in ER isolated from PE & PTC placentae (PE & PTC, n = 3 separate samples). c WB & associated densitometry of ASAH2 (normalized to TOM20) in mitochondria from PTC & PE placentae. d Representative TEM depicting mitophagy (white arrow) in cytotrophoblast cell from PE placenta (scale bar: 500 nm, n = 8 separate PE placentae). e Western blot & associated densitometry of PINK1 & Parkin in PE vs. PTC mitochondrial isolates. Densitometry for PINK1 blot was used to calculate the ratio of full-length PINK63kDa to cleaved PINK153kDa (PE & PTC, n = 4 separate placentae, *P < 0.05) Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41419-018-0360-0), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Deficiency or mutant PINK1 reduces phosphorylation of LETM1 at Thr192. a, b Proteins extracted from PINK1 WT or KO MEFs (a) or mouse brain (b) were subjected to IP with anti-LETM1, probed with pT192 & reprobed with anti-LETM1 by WB. c Proteins extracted from human fibroblast of control (Con) or a PINK1-Q456X patient (Q456X) were subjected to IP with anti-LETM1, probed with pT192 & reprobed with anti-LETM1 by WB. d HEK293 cells were transfected with Adtrack GFP control, AdPINK1-WT, & AdPINK1-Q456X mutant for 1 day. Endogenous LETM1 protein was isolated by IP with anti-LETM1, probed with pT192 & reprobed with anti-LETM1 by WB. e SH-SY5Y cells were treated with 25 µM rotenone for 8 or 16 h. Total cell lysates were either analyzed by WB with anti-PINK1, or subjected to IP with anti-LETM1, probed with pT192 & reprobed with anti-LETM1 antibodies by WB. All above experiments were replicated three times, respectively Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/29123128), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Polyphyllin I suppressed, & PINK1 knockdown further suppressed, tumor growth in a MDA-MB-231 xenograft modelA. Tumor volumes were measured every week & differed at the end of the treatment period (*P < 0.01 compared to shCon cells treated with PPI, ##P< 0.01 compared to shPINK1 cells treated with PPI). B. Representative image of tumors from each group. C. Body weight changes in mice during the 6 weeks of PPI treatment. There were no differences in body weights between the PPI-treated shCon & vehicle-treated shCon groups or between the PPI-treated shPINK1 group & shCon groups. D. Representative tumor tissues were sectioned & subjected to H&E staining, TUNEL assay, & immunohistochemistry staining for C-CASP3. Scale bars: 50 μm. E. Representative tumor tissues from each group were prepared & subjected to western blot using anti-DRP1, -PARK2, -LC3B, -PINK1, & -C-CASP3 antibodies. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.14413), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Corticosterone affects NIX-dependent mitophagy through decreasing PGC1 alpha in vivo.a–f Mice exposed to vehicle, corticosterone (10 mg/kg), corticosterone w/ phorbol 12-myristate 13-acetate (PMA pretreatment, 200 μg/kg), or PMA alone for 7 days. a Slide samples for IHC immunostained w/ LAMP1 (green), TOMM20 (red), & DAPI (blue). Scale bars, 100 μm (magnification, ×200). n = 5. b The expressions of NIX, PTEN-induced kinase 1 (PINK1), & BCL2 interacting protein 3 (BNIP3) detected w/ WB where beta-actin used as a loading control. n = 5. c Slide samples for IHC immunostained w/ synpatophysin (green), PSD95 (red), & DAPI (blue). Scale bars, 100 μm (magnification, ×200). n = 5. d Synaptophysin & PSD95 detected by WB. Loading control is beta-actin. n = 5. e The mice subjected to Y-maze test to evaluate spatial memory function. n = 6. f The mice subjected to forced swim test to evaluate depression-like behavior. n = 5. g Vehicle or RU 486 (5 mg/kg) injected mice presented w//w/out corticosterone (10 mg/kg) for 3 days. The expressions of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1 alpha) & NIX visualized via WB. Loading control is beta-actin. n = 5. h The schematic model for mechanisms of inhibition in NIX-dependent mitophagy by glucocorticoid shown. All blots & IF images representative. n = 5 or 6 from each animal w/ two technical replicates each in results of IHC & WB. Quantitative data presented as a mean ± S.E.M. The representative images acquired by SRRF imaging system. Two-sided two-way ANOVA conducted except Fig. 8b, data of which analyzed by two-sided unpaired student’s t test. ** indicates p < 0.01 versus control & ## indicates p < 0.01 versus corticosterone, respectively. Data provided as a Source data file. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/33473105), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - G-TPP activity is conserved in primary fibroblasts(A, C) Fibroblasts were treated with 15 µM G-TPP for the indicated time points. Cells were harvested & western blots were probed with antibodies against (A) PINK1, pS65-Ub & total Ub or (C) autophagy adapter proteins. GAPDH & Vinculin served as loading control. G-TPP treatment led to PINK1 stabilization & pS65-Ub induction in primary skin fibroblasts. p62 levels were induced upon G-TPP treatment, while other adapters seemed decreased. (B, D) Human fibroblasts were treated with 15 µM G-TPP for 16 h & fixed & stained with antibodies against (B) pS65-Ub (green) or (D) the autophagy adapters NBR1, NDP52, p62, OPTN & TAX1BP1 (green). Mitochondria were stained with antibodies against TOM20 (red), nuclei were visualized with Hoechst (blue). Scale bars indicate 10 µM. A magnified image of the boxed region, the fluorescence profile along the arrow & the Pearson’s correlation coefficient of adapter protein & mitochondrial stainingare shown to the right. Shown is the mean ± SEM of at least five randomly selected images (unpaired, two-sided t-test, ***p < 0.0005). Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.22287), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Polyphyllin I triggers PINK1/PARK2-dependent mitophagyA. MDA-MB-231 cells were treated with 8 μM PPI for different periods of time as indicated, & PARK2, P62, & ubiquitin (UB) levels in mitochondrial fractions were determined by western blot. B. Cells were cotransfected with GFP-UB & RFP-mito & treated with 8 μM PPI for 9 h, after which PARK2 (Alexa Fluor 405, pink) & P62 (Alexa Fluor 647, blue) immunostaining was detected using confocal microscopy. C. Cells cotransfected with GFP-UB & RFP-LC3 were treated with 8 μM PPI for 9 h, after which PARK2 (Alexa Fluor 405, pink) & TOMM20 (Alexa Fluor 647, blue) immunostaining was detected using confocal microscopy. Scale bars: 10 μm. D-E. MDA-MB-231 cells were treated with 8 μM PPI for different periods of time as indicated; whole-cell lysates were then separated on 8% SDS-PAGE gels & analyzed by western blot using the anti-PINK1 antibody. Relative full-length (∼63 kDa) & cleaved (∼52 kDa) PINK1 levels were quantified by densitometry & normalized to Tubulin. The results were expressed as a percentage of control, which was set at 100%. Data are presented as mean ± SD (*P< 0.01 vs. the control). F. Cells were treated with 8 μM PPI for different periods of time as indicated, & whole-cell lysates were then subjected to western blot analysis. G. Cells were treated with 8 μM PPI for 9 h, after which mitochondrial fractions were prepared & subjected to immunoprecipitation using anti-PINK1 antibody; associated PARK2 was detected using immunoblotting. H. RFP-mito-expressing MDA-MB-231 cells were treated with 8 μM PPI for 9 h, & PINK1 (Alexa Fluor 488, green) & PARK2 (Alexa Fluor 405, pink) immunostaining were evaluated using confocal microscopy. Scale bars: 10 μm. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.14413), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - miR-27a/b suppress human PINK1 expression. a Schematic diagram of the conserved target sites of miR-27a/b in the 3′UTR of human PINK1 mRNA. Seed sequences were indicated in red. Asterisks (*) indicate the conserved nucleotides. b Schematic diagram of the luciferase reporter plasmids. The reporter constructs contain the full-length 3′UTR of human PINK1 mRNA with wild-type (WT) or mutated (Mut) seed match sites downstream of Renilla luciferase gene. c Overexpression of miR-27a/b decreased luciferase activities in HeLa cells. Cells were transfected with miR-27a/b or negative control (Ctl) along with reporter constructs as indicated. 48 h post-transfection, luciferase activities were measured (n = 4, one-way ANOVA). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity. d-g Overexpression of miR-27a/b decreased PINK1 protein levels in HeLa (D-E) & M17 cells (F-G). Cells were transfected with miR-27a/b or negative control (Ctl). 48 h post-transfection, cells were harvested for Western blot (n = 4, t-test). PINK1 protein level was normalized to corresponding GAPDH level & quantified as a percentage of control. Data are shown as a percentage of control. Values are mean ± SEM (n.s. = non-significant, **p < 0.01, ***p < 0.001) Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/27456084), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - Increased p62 expression is prevented in PINK1 knock‐down cells. (a) SH‐SY5Y cells expressing scrambled shRNA or PINK1 shRNA (117‐2, 118‐3, & 118‐4) were treated with 10 μM carbonyl cyanide m‐chlorophenylhydrazone (CCCP) for 24 h & full‐length (FL) PINK1 protein levels measured by western blot. No accumulation of FL PINK1 was observed in 118‐3 & 118‐4 PINK1 shRNA clones. (b) SH‐SY5Y cells constitutively expressing scrambled (scram) or PINK1 shRNA (clone 118‐3) were treated with vehicle or 10 μM CCCP for 24 h & p62 protein expression measured by western blotting. **p < 0.05 versus scram CCCP; n = 9. (c) SH‐SY5Y cells were incubated in serum‐free culture media for up to 24 h to induce starvation‐mediated macroautophagy. p62 protein levels were measured by western blotting. *p < 0.05 versus 0 h; **p < 0.01 versus 0 h; n = 4. (d) SH‐SY5Y cells were transfected with scrambled (scram) or p62 siRNA for 72 h. For the last 24 h, cells were treated with vehicle (ethanol) or 10 μM CCCP. p62 protein expression was then measured by western blotting. p62 protein was significantly decreased in p62 siRNA cells under basal conditions & following CCCP treatment, when compared to scram cells. **p < 0.01 versus scram vehicle or scram CCCP; n = 3. (e) Mitochondrial content was measured in SH‐SY5Y cells treated with scram or p62 siRNA for 72 h by measuring citrate synthase (CS) activity. For the last 24 h, cells were treated with vehicle or 10 μM CCCP & data are expressed as % of vehicle. *p < 0.05 versus scram CCCP treatment; n = 5. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/26509433), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - NF kappaB upregulates human PINK1 gene expression. (A) NF kappaB increases the endogenous human PINK1 mRNA level. HEK293 cells were transfected with either the NF kappaB expression vector or empty vector pMTF. RT-PCRs were performed using either primers specific to the human PINK1 coding sequence or the human beta-actin coding sequence. (B) NF kappaB increases the endogenous human PINK1 protein levels. HEK293 cells transfected with NF kappaB were harvested 48 h after transfection for protein detection. Cell lysates were run on 10% glycine gel & images were collected by Licor. A significant increase of endogenous PINK1 was observed. (C) The endogenous human PINK1 protein level was dramatically increased by NF kappaB in SH-SY5Y cells. SH-SY5Y cells were transfected with NF kappaB p65 plasmids or the control vector pMTF & then harvested for protein detections. The human PINK1 protein & its proteolysis products ∆1-PINK1 (55kD) & ∆2-PINK1 (45kD) were all increased. beta-actin acted as the internal control. (D) LPS treatment facilitated PINK1 expression in SH-SY5Y cells. Cells were harvested after being treated for 16 h & then subjected to Western blot. The human beta-actin level was served as a control. Quantification was performed by Image J software. Values indicate means ± SEM. n = 3, *p <0.01 by Student’s t-test. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25108683), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - G-TPP induces PINK1 stabilization & kinase activity in HeLa cells(A) G-TPP treatment leads to PINK1 stabilization & pS65-Ub induction in HeLa cells. HeLa cells stably expressing untagged Parkin were treated with 10 µM G-TPP for the indicated times. Western blots were prepared with cell lysates & probed with antibodies against PINK1 & pS65-Ub. GAPDH served as a loading control. (B) pS65-Ub is induced in G-TPP treated cells & co-localizes with EGFP-Parkin & mitochondria. HeLa cells stably expressing EGFP-Parkin (green) were treated with 10 µM G-TPP for the indicated times & fixed. Cells were stained with antibodies against pS65-Ub (red) & the mitochondrial marker TOM20 (cyan). Scale bars correspond to 10 µM. (C) Quantification of Parkin translocation using High Content Imaging. HeLa EGFP-Parkin cells were treated for 4 or 8 h with or without 10 µM G-TPP. CCCP treatment (10 µM for 2 h) was used as a positive control. Cells were fixed, counterstained with Hoechst dye to visualize nuclei, imaged & analyzed using the ratio of cytoplasmic to nuclear EGFP signal [21]. Data was normalized to positive (2 h 10 µM CCCP treatment) & negative (2 h DMSO) controls. G-TPP significantly induced Parkin re-localization to levels similar to or beyond 2 h CCCP treatment. Shown are the mean values of three independent experiments with triplicate wells each ± SEM (one-way ANOVA with Tukey’s posthoc, ***p < 0.0005). Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.22287), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - NF kappaB upregulates human PINK1 gene expression. (A) NF kappaB increases the endogenous human PINK1 mRNA level. HEK293 cells were transfected with either the NF kappaB expression vector or empty vector pMTF. RT-PCRs were performed using either primers specific to the human PINK1 coding sequence or the human beta-actin coding sequence. (B) NF kappaB increases the endogenous human PINK1 protein levels. HEK293 cells transfected with NF kappaB were harvested 48 h after transfection for protein detection. Cell lysates were run on 10% glycine gel & images were collected by Licor. A significant increase of endogenous PINK1 was observed. (C) The endogenous human PINK1 protein level was dramatically increased by NF kappaB in SH-SY5Y cells. SH-SY5Y cells were transfected with NF kappaB p65 plasmids or the control vector pMTF & then harvested for protein detections. The human PINK1 protein & its proteolysis products ∆1-PINK1 (55kD) & ∆2-PINK1 (45kD) were all increased. beta-actin acted as the internal control. (D) LPS treatment facilitated PINK1 expression in SH-SY5Y cells. Cells were harvested after being treated for 16 h & then subjected to Western blot. The human beta-actin level was served as a control. Quantification was performed by Image J software. Values indicate means ± SEM. n = 3, *p <0.01 by Student’s t-test. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25108683), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - NF kappaB upregulates human PINK1 gene expression. (A) NF kappaB increases the endogenous human PINK1 mRNA level. HEK293 cells were transfected with either the NF kappaB expression vector or empty vector pMTF. RT-PCRs were performed using either primers specific to the human PINK1 coding sequence or the human beta-actin coding sequence. (B) NF kappaB increases the endogenous human PINK1 protein levels. HEK293 cells transfected with NF kappaB were harvested 48 h after transfection for protein detection. Cell lysates were run on 10% glycine gel & images were collected by Licor. A significant increase of endogenous PINK1 was observed. (C) The endogenous human PINK1 protein level was dramatically increased by NF kappaB in SH-SY5Y cells. SH-SY5Y cells were transfected with NF kappaB p65 plasmids or the control vector pMTF & then harvested for protein detections. The human PINK1 protein & its proteolysis products ∆1-PINK1 (55kD) & ∆2-PINK1 (45kD) were all increased. beta-actin acted as the internal control. (D) LPS treatment facilitated PINK1 expression in SH-SY5Y cells. Cells were harvested after being treated for 16 h & then subjected to Western blot. The human beta-actin level was served as a control. Quantification was performed by Image J software. Values indicate means ± SEM. n = 3, *p <0.01 by Student’s t-test. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25108683), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: PINK1 Antibody - BSA Free [BC100-494] -
Western Blot: PINK1 Antibody - BSA Free [BC100-494] - G-TPP leads to recruitment of autophagy adapters & degradation of mitochondria(A) HeLa cells stably expressing untagged Parkin were treated with 10 µM G-TPP for 8 h. Western blots were prepared from cell lysates & probed with antibodies against LC3, phospho-TBK1 (Ser172) & TBK1. GAPDH was used as a loading control. Upon 8 h the levels of LC3-I & LC3-II were both increased. At 8 h after treatment with G-TPP but not at 4 or 24 h, TBK1 was phosphorylated. (B) HeLa cells stably expressing EGFP-Parkin were treated with 10 µM G-TPP & fixed 8 h after treatment. Cells were stained with antibodies against the autophagy adapter proteins NBR1, NDP52, OPTN, p62, & TAX1BP1 (red). Mitochondria were counterstained with TOM20 antibodies (cyan), nuclei with Hoechst (blue). EGFP-Parkin epifluorescence is shown in green. Scale bar corresponds to 10 µM. (C) HeLa cells stably expressing EGFP-Parkin & the reporter protein mitoKeima were treated with 10 µM CCCP or G-TPP & imaged over time. The ratio of ‘neutral’ mitoKeima to ‘acidic’ mitoKeima was calculated as readout for mitophagy. Parkin translocation was monitored at the same time. Values for Parkin translocation & mitophagy were normalized to 12 h treatment with 10 µM CCCP as positive control & DMSO as negative control (two-way ANOVA with Tukey’s post-hoc test, **p < 0.005, ***p < 0.0005). Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.22287), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: Rabbit Polyclonal PINK1 Antibody [BC100-494]
Western Blot: Rabbit Polyclonal PINK1 Antibody [BC100-494] - PINK1 detection in mouse and postmortem human cortex samples. 1:1000 dilution at 4 degrees overnight to detect endogenous PINK1 from mouse and postmortem human cortex samples (10 ug total protein). Image from a verified customer review.












![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-271220231253319.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241534312.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415384188.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241534386.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241538244.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241534536.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415334919.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415363734.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-3102024153976.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241539713.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-3102024153979.jpg)
![Immunocytochemistry/ Immunofluorescence: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241533498.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482291.jpg)
![Immunocytochemistry/ Immunofluorescence: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482253.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482226.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415483624.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415485127.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241548513.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415483642.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415485119.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415483643.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482289.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241548362.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482229.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415483619.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482287.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482296.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415485118.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482279.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482299.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415483648.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482218.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482286.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482214.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415483618.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482278.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415485116.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241548369.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482277.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-31020241548363.jpg)
![Western Blot: PINK1 Antibody - BSA Free [BC100-494] - PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/bc100-494_rabbit-polyclonal-pink1-antibody-310202415482225.jpg)
![Western Blot: Rabbit Polyclonal PINK1 Antibody [BC100-494] PINK1 Antibody - BSA Free](https://resources.bio-techne.com/images/products/antibody/bc100-494_rabbit-polyclonal-pink1-antibody-western-blot-231202510057..jpg)