Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130]
Immunohistochemistry-Paraffin: TRF-2 Antibody [NB110-57130] - Analysis in xenografted human breast cancer tissue using DAB with hematoxylin counterstain.
Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130]
Immunohistochemistry-Paraffin: TRF-2 Antibody [NB110-57130] - Analysis of FFPE human breast cancer tissue with rabbit polyclonal TRF2 antibody at a dilution of 1:200. The staining was developed with HRP-DAB detection method and the counterstaining was performed using hematoxylin. This TRF2 antibody generated an expected nuclear signal in all the cancer cells and the stromal cells. In the tested section, only a subset of myoepithelial cells showed positivity for this protein.
Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130]
Immunocytochemistry/Immunofluorescence: TRF-2 Antibody [NB110-57130] - NIH3T3 cells were fixed in 4% paraformaldehyde for 10 minutes and permeabilized in 0.5% Triton X-100 in PBS for 5 minutes. The cells were incubated with anti-TRF-2 Antibody NB110-57130 at 2 ug/ml overnight at 4C and detected with an anti-rabbit Dylight 488 (Green) at a 1:1000 dilution for 60 minutes. Nuclei were counterstained with DAPI (Blue). Cells were imaged using a 100X objective and digitally deconvolved.
Flow Cytometry: TRF-2 Antibody - BSA Free [NB110-57130]
Flow Cytometry: TRF-2 Antibody [NB110-57130] - An intracellular stain was performed on HeLa cells with TRF-2 Antibody NB110-57130 (blue) and a matched isotype control NBP2-24891 (orange). Cells were fixed with 4% PFA and then permeabilized with 0.1% saponin. Cells were incubated in an antibody dilution of 1.0 ug/mL for 30 minutes at room temperature, followed by Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Dylight 550 (SA5-10033, Thermo Fisher).
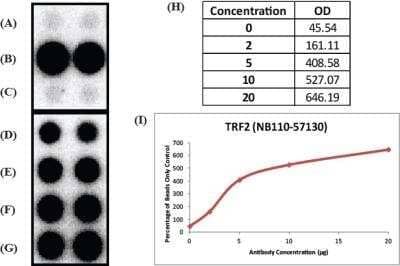
Chromatin Immunoprecipitation: TRF-2 Antibody [NB110-57130] - Analysis in mouse. Titrated TRF2 antibody to determine concentration required for ChIP experiment. ChIP image submitted by a verified customer review.
Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130]
Immunocytochemistry/Immunofluorescence: TRF-2 Antibody [NB110-57130] - RNAi-mediated depletion of human separase (ESPL1) induces TIFs. Control scrambled siRNA- (control) and ESPL1 siRNA-treated fibroblasts stained with anti-p53-binding protein 1 (53BP1; green) and anti-TRF2 (red). It is noteworthy that in ESPL1 siRNA-treated cells, 53BP1 signals frequently overlap with TRF2 signals marking the TIFs. Scale bar, 5 um. Image collected and cropped by CiteAb from the following publication (https://www.nature.com/doifinder/10.1038/ncomms10405), licensed under a CC-BY license.
Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130]
Immunohistochemistry-Paraffin: TRF-2 Antibody [NB110-57130] - Analysis of FFPE human breast cancer tissue with rabbit polyclonal TRF2 antibody at 1:200 dilution. The staining was developed with HRP-DAB detection method and the counterstaining was performed using hematoxylin. This TRF2 antibody generated an expected nuclear signal in all the cancer cells and the stromal cells. In the tested section, only a subset of myoepithelial cells showed positivity for this protein.
Flow Cytometry: TRF-2 Antibody - BSA Free [NB110-57130]
Flow Cytometry: TRF-2 Antibody [NB110-57130] - An intracellular stain was performed on HeLa cells with TRF-2 Antibody NB110-57130AF488 (blue) and a matched isotype control (orange). Cells were fixed with 4% PFA and then permeabilized with 0.1% saponin. Cells were incubated in an antibody dilution of 5 ug/mL for 30 minutes at room temperature. Both antibodies were conjugated to Alexa Fluor 488.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
The DNA damage response at telomeres uncapped through TRF2 depletion does not require BRCA1 or CtIPImmortalized Brca1F/− MEFs were infected with retroviruses expressing the indicated shRNAs and/or Cre recombinase, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later & analysed by Western blotting as indicated. SMC1 & tubulin were used as loading controls. *non-specific band.Cells treated as in (A) were fixed 48 h after selection & stained with an anti-53BP1 antibody (green). Telomeres were visualized with a Cy3-conjugated (CCCTAA)3-PNA probe (red). Yellow arrowheads point to 53BP1 foci that co-localize with telomeres.Quantification of TIFs in cells treated as in (B). A minimum of 200 nuclei were scored for each sample. Error bars represent SD of two independent experiments. P-values were calculated using an unpaired two-tailed t-test. *P ≤ 0.05; NS, P > 0.05. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25582120), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Repressive chromatin induced on p21 promoter in TRF2 dependent way. (A–C) Loss of chromatin activation marks H3K4Me, H3K4Me2 in stable TRF2 expressed HT1080 cells (A); increase in LSD1 occupancy in TRF2 over-expressing HT1080 cells. (B) (Full images are shown in Supplementary Figures S6 & S7); Co-immunoprecipitation of TRF2 with LSD1 (immunoprecipitation with anti-TRF2 antibody followed by immunoblotting with anti-LSD1 or anti-TRF2 antibody) (C); (D) Reverse Co-immunoprecipitation of LSD1 with TRF2 (immunoprecipitation with anti-LSD1 antibody followed by immunoblotting with anti-TRF2 antibody); (E) & reduced occupancy of the repressor complex REST, Co-REST & LSD1 on silencing TRF2 on p21 promoter. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-11177-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] -
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 occupancy on G4-motif sites on gene promoters within cells.A, telomeric enrichment was tested for TRF2 ChIP samples performed in HT1080 cells. Telomeric signal was normalized to signal from ALU probe. B, TRF2 occupancy at gene promoter sites in HT1080 cells (endogenous TRF2 & TRF2-overexpressed) quantified using ChIP-qRT PCR. TRF2 ChIP/IgG (mock) enrichment was normalized to 1% input; CTCF promoter (which does not harbor a TRF2 peak) was used as negative control; error bars correspond to S.D., & statistical significance was calculated by paired t test (*, p < 0.05; **, p < 0.01). For the TRF2-overexpressed condition (in red), all p values were <0.01. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31575660), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Repressive chromatin induced on p21 promoter in TRF2 dependent way. (A–C) Loss of chromatin activation marks H3K4Me, H3K4Me2 in stable TRF2 expressed HT1080 cells (A); increase in LSD1 occupancy in TRF2 over-expressing HT1080 cells. (B) (Full images are shown in Supplementary Figures S6 & S7); Co-immunoprecipitation of TRF2 with LSD1 (immunoprecipitation with anti-TRF2 antibody followed by immunoblotting with anti-LSD1 or anti-TRF2 antibody) (C); (D) Reverse Co-immunoprecipitation of LSD1 with TRF2 (immunoprecipitation with anti-LSD1 antibody followed by immunoblotting with anti-TRF2 antibody); (E) & reduced occupancy of the repressor complex REST, Co-REST & LSD1 on silencing TRF2 on p21 promoter. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-11177-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] -
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - Thousands of extra-telomeric TRF2 binding sites found across the genome.A, extra-telomeric TRF2 peaks found in HT1080 cells following TRF2 ChIP-Seq: 20304 TRF2 peaks were common between two independent experiments. B, distribution of common TRF2 peaks around TSSs. Distance from the TSS is shown in kb. C, replicate consistency plot generated using irreproducible discovery rate analysis; 1956 peaks found at ≤0.01 are marked. D, distribution of 1956 TRF2 peaks with IDR 0.01 (TRF2HC peaks) around the TSS. Distance from the TSS is shown in kb. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31575660), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF1HP1 alpha allele-specific protection effects upon si-TRF2-induced telomeric damage. 72 h after transfection, a TRF2 knockdown efficiency with antibody against TRF2 (anti-TRF2) & GAPDH (anti-GAPDH) as loading control. (−) si-non-targeting; (+) si-TRF2. Quantification of TIFs in b si-non-targeting (n = 32–47 nuclei per group) or c si-TRF2. Left *p = 0.0188, right *p = 0.0192, **p = 0.0042, ****p < 0.0001 (n = 31–48 nuclei per group). b, c Significance is assessed by one-way ANOVA & Dunnett’s multiple comparison test with 95% confidence level. Error bars represent s.e.m. Note the similar pattern among TRF1HP1 alpha alleles in c compared to the corresponding allele pattern in Fig. 4b Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/30181605), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] -
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 peaks harbor G-quadruplex motifs genome-wide.A, schematic representation of a G4 motif; sequence pattern with loop/stem & PG4 motif formed by a tetrad of guanine trimers interspersed with loops that can vary in length. B & C, PG4 motifs & TRF2 peaks significantly overlap. High-confidence TRF2-binding sites (TRF2HC peaks) determined by ChIP-Seq in HT1080 cells were significantly enriched in PG4-motif sequences (B), & conversely, PG4-motif sequences were enriched within TRF2HC peaks (C). Nonoverlapping PG4 motifs were considered for analysis; for control analysis, 100 regions of identical length for each TRF2 peak were taken. *, p < 0.05; **, p < 0.01 (Fisher's exact test). Error bars, S.D. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31575660), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Repressive chromatin induced on p21 promoter in TRF2 dependent way. (A–C) Loss of chromatin activation marks H3K4Me, H3K4Me2 in stable TRF2 expressed HT1080 cells (A); increase in LSD1 occupancy in TRF2 over-expressing HT1080 cells. (B) (Full images are shown in Supplementary Figures S6 & S7); Co-immunoprecipitation of TRF2 with LSD1 (immunoprecipitation with anti-TRF2 antibody followed by immunoblotting with anti-LSD1 or anti-TRF2 antibody) (C); (D) Reverse Co-immunoprecipitation of LSD1 with TRF2 (immunoprecipitation with anti-LSD1 antibody followed by immunoblotting with anti-TRF2 antibody); (E) & reduced occupancy of the repressor complex REST, Co-REST & LSD1 on silencing TRF2 on p21 promoter. Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-11177-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 transcriptionally regulates p21 expression through promoter occupancy. (A) Quantitative ChIP using TRF2 antibody gives enriched binding of TRF2 on p21 promoter in HT1080 & MDA-MB-231 cells, IgG was used as isotypic antibody control; normalized with 10% input (data represented as mean ± SEM, for three replicates). (B,C) TRF2 represses p21 promoter activity. In luciferase assay, si-RNA-mediated silencing or TRF2 over expression resulted in increased (B, *p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) or reduction (C, *p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) in p21 promoter activity in HT1080 & MDA-MB-231 cells, respectively; over expression of TRF2 devoid of DNA binding [deletion of basic (delB), myb (delM) & both basic/myb (delB-delM) domains] resulted in partial or complete rescue of p21 promoter activity in HT1080 & MDA-MB-231 cells (data represented as mean ± SEM, for three replicates) (C). (D,E) HT1080 cells over expressing TRF2 show reduced p21 protein expression, bar graph shows the densitometry analysis of three different immunoblot replicates (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates). (E) (Full images are shown in Supplementary Figure S5); TRF2 silencing results in increase in p21 protein, bar graph shows the densitometry analysis of three different immunoblots (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) (F); & mRNA expression in HT1080 cells (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates, GAPDH used as internal control for real-time PCR). Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-11177-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] -
Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 occupancy at gene promoters is sensitive to intracellular G-quadruplex–binding ligand 360A.A, TRF2 occupancy at gene promoter sites was checked by ChIP qRT-PCR in HT1080 cells following overexpression of TRF2 in the presence or absence of 360A. Error bars, S.D. from three independent experiments; CTCF promoter was used as a negative control. B, TRF2 level was checked by Western blotting in the presence of the ligand 360A in both untransfected & TRF2 transient overexpression conditions. Overexpression was also confirmed by probing for DDK tag. GAPDH was used as loading control. C, expression of target genes was analyzed by qRT-PCR in HT1080 cells following overexpression of TRF2 in the presence or absence of 360A. GAPDH expression was used for normalization; error bars, S.D. from three independent experiments. Statistical significance was calculated by paired t test (*, p < 0.05; **, p < 0.01). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31575660), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - A. Co-IP analysis of the interaction between DSP & TRF2 in HEK293 cells & MCF-7 cells. B. Co-IP analysis of the interaction between DSP & TRF2 in DSP-EGFP transfected HEK293 cells or DSP△NLS-EGFP transfected HEK293 cells. C. DSP pull down by telomere targeting CASID with the condition of TRF2 knockdown in HEK293 cells. Histone H3 is the loading control. D.TRF1 & TRF2 pull-down assay by telomere targeting CASID with the condition of DSP knockdown in HEK293 cells. E. Left, Dot blot of telomere DNA repeats in chromatin pull down by DSP with the condition of TRF1 or TRF2 knockdown. The dot-blot was performed at least three times & the represented one is shown. Right, the histogram shows the quantitated results of the dot blots. The triple asterisks (***) indicates a p-value less than 0.01. ns. indicates the insignificance difference. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31595153), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] -
Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] - TERRA & telomere transcription in ALT cells.(a) TERRA northern blot hybridizations of RNA from the indicated cell lines (VA13: WI-38 VA13; 1.2.11: HeLa 1.2.11) pre-treated with RNaseA or left untreated. Ethidium bromide stained 18S ribosomal RNA (rRNA) is shown to control for loading. Long TERRA molecules comprised between the wells of the gels (w) & 28S rRNA are indicated. (b) TERRA CpG-island promoter methylation analysis of the indicated cell lines. Genomic DNA was digested with the methylation sensitive restriction enzyme MspI or its methylation insensitive isoschizomer HpaII. DNA was hybridized using a radioactively labelled probe detecting TERRA promoter CpG-island repeats. Nomet: fragments corresponding to unmethylated restriction products. (c) Dot blot hybridization of DNA immunoprecipitated with antibodies against phosphorylated Serines S2 & S5 of RNA polymerase II C-terminal domain. Hybridizations were performed with a telomeric probe. Quantifications are shown at the bottom. (d) Bars & error bars are averages & s.d. from three independent experiments. (e) Examples of TERRA FISH in the indicated cells. TERRA is shown in red, DAPI-stained DNA in blue. Scale bar, 9 μm. (f) IF/FISH experiments in the indicated cell lines. TERRA is in red, TRF2 in green & PML in blue. In the merge panels, arrowheads point to nuclear foci where the three factors co-localize. Scale bar, 9 μm. (g) Information surface at 0.01 μm detail level of three TERRA-containing APBs. TERRA is in red, TRF2 in green & PML in cyan. Images were generated with Three-Dimensional Structured Illumination Microscopy (3D-SIM). Scale bars, 0.4 μm. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25330849), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 occupancy at gene promoters is sensitive to intracellular G-quadruplex–binding ligand 360A.A, TRF2 occupancy at gene promoter sites was checked by ChIP qRT-PCR in HT1080 cells following overexpression of TRF2 in the presence or absence of 360A. Error bars, S.D. from three independent experiments; CTCF promoter was used as a negative control. B, TRF2 level was checked by Western blotting in the presence of the ligand 360A in both untransfected & TRF2 transient overexpression conditions. Overexpression was also confirmed by probing for DDK tag. GAPDH was used as loading control. C, expression of target genes was analyzed by qRT-PCR in HT1080 cells following overexpression of TRF2 in the presence or absence of 360A. GAPDH expression was used for normalization; error bars, S.D. from three independent experiments. Statistical significance was calculated by paired t test (*, p < 0.05; **, p < 0.01). Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31575660), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - A. Gene ontology analysis of proteins identified by CASID targeting telomeres. B. Western blot detection of TRF1, POT1, & DSP in the proteins pulled down by CASID with the presence of telo-sgRNA in HEK293 cells. Input represents the initial nuclear extract before the pull-down mediated by CASID. C. left, ChIP assay of telomere binding of TRF2 or DSP in MCF-7 cells & HEK293 cells. The chromatin DNA precipitated by anti-DSP & anti-TRF2 antibodies were analyzed by dot blot using the probes recognizing telomere (Telo-p) or Alu DNA repeats (Alu-p). The detection of Alu DNA repeats serves as negative control. The dot-blot was performed at least three times & the represented one is shown. Right, the histogram shows the quantitated results of the dot blots. The triple asterisks (***) indicates a p-value less than 0.01. D. Fluorescence imaging of DSP-EGFP in MCF-7 cells & U2OS cells. The EGFP signal indicates the localization of DSP-EGFP (green) & the Telo-FISH signal indicates the localization of telomeres (red). The overlapping between DSP-EGFP & Telo-FISH is in yellow as revealed in the enlarged nuclear area. The dCAS9-EGFP construct without telo-sgRNA was used for U2OS transfection & the imaging of EGFP serves as negative control. E. DSP localization in interphase & pro-metaphase nucleus. DSP-EGFP indicates the DSP locations & DAPI indicates the nucleus. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31595153), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 interacts with lamin A/C.(a) Pull down of endogenous TRF2 in IMR90s. The top panel shows a blot probed for endogenous lamin A/C, & the bottom panel shows a blot with the same samples probed for TRF2. (b) Pull down of endogenously expressed lamin A/C in IMR90s. The top panel shows a blot probed for endogenous TRF2, & the bottom panel shows a blot with the same samples probed for endogenous lamin A/C. (c) Pull down of endogenous lamin A/C in IMR90s exogenously expressing GFP, GFP-TRF2, GFP-TRF deltaB deltaM or GFP-TRF1. The top panel shows a blot probed for GFP, & the bottom panel shows the same blot probed for lamin A/C. (d) Pull down of endogenous TRF2 in IMR90s exogenously expressing GFP, GFP-LMNA or progerin (GFP-LMNA delta50). The top panel shows a blot probed for GFP, & the bottom panel shows the same blot probed for TRF2. Asterisks denote IgG bands. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25399868), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Widespread dysregulation of shelterin protein expression in bortezomib-treated HEL & BGC-823 cellsA. & B. mRNA levels of shelterin factors TRF1, TRF2, TPP1, POT1, RAP1 & TIN2 in bortezomib-treated cells. Cells were treated with bortezomib for 24 hours & qPCR was used for quantitative assays. The levels of each target mRNA in bortezomib-treated cells were expressed as percentages of those in untreated cells. (A) HEL cells & (B) BGC-823 cells. C. & D. Immunoblotting assessment of TRF1, TRF2 & POT1 protein expression in bortezomib-treated cells. Same sets of cells above were analyzed for TRF1, TRF2 & POT1 protein levels & shown was representative of three independent experiments. (C) HEL cells & (D) BGC-823 cells. * & **: P < 0.05 & 0.01, respectively. BTZ, bortezomib. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.5752), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - A. Co-IP analysis of the interaction between DSP & TRF2 in HEK293 cells & MCF-7 cells. B. Co-IP analysis of the interaction between DSP & TRF2 in DSP-EGFP transfected HEK293 cells or DSP△NLS-EGFP transfected HEK293 cells. C. DSP pull down by telomere targeting CASID with the condition of TRF2 knockdown in HEK293 cells. Histone H3 is the loading control. D.TRF1 & TRF2 pull-down assay by telomere targeting CASID with the condition of DSP knockdown in HEK293 cells. E. Left, Dot blot of telomere DNA repeats in chromatin pull down by DSP with the condition of TRF1 or TRF2 knockdown. The dot-blot was performed at least three times & the represented one is shown. Right, the histogram shows the quantitated results of the dot blots. The triple asterisks (***) indicates a p-value less than 0.01. ns. indicates the insignificance difference. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31595153), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Telomere deprotection contributes to replication stress lethality. a Western blots of whole cell extracts from HT1080 6TG cells stably transduced with control, TRF2 shRNA (TRF sh-F) or TRF2 over expression (TRF2OE) vectors. b Representative images of cyto-centrifuged chromosome spreads stained with DAPI (blue), gamma-H2AX IF (red) & telomere PNA (green) from control, TRF sh-F or TRF2OE HT1080 6TG cells treated with DMSO or APH. Scale bar represents 10 µm. c, d Quantitation of mitotic telomere DDR foci from Control sh & TRF2 sh-F cells (c) or vector & TRF2OE cells (d) ± DMSO or APH (three biological replicates scoring n = 50 mitotic spreads per replicate compiled in a Tukey box plot, Mann–Whitney test). e Difference in the mean number of mitotic telomeric gamma-H2AX foci between HT1080 6TG TRF2 sh-F or TRF2OE cells & their appropriate vector control. These are a different representation of the same data shown in (c, d) (mean ± s.e.m., n = 3 three biological replications, Student’s t-test). f Mitotic duration to cell death in APH treated control, TRF2 sh-F & or TRF2OE cells (three biological replicates scoring ≥267 mitotic death events per condition are shown in a dot plot, mean ± s.e.m., Mann–Whitney test). g Mitotic outcome of control, TRF2 sh-F & TRF2OE cells treated with APH or DMSO (mean ± s.e.m., n = 3 biological replicates of at least 267 mitoses per condition, Fisher’s Exact Test). For all panels, *p < 0.05, **p < 0.01. Source data are provided as a Source Data file Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31530811), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - A. Co-IP analysis of the interaction between DSP & TRF2 in HEK293 cells & MCF-7 cells. B. Co-IP analysis of the interaction between DSP & TRF2 in DSP-EGFP transfected HEK293 cells or DSP△NLS-EGFP transfected HEK293 cells. C. DSP pull down by telomere targeting CASID with the condition of TRF2 knockdown in HEK293 cells. Histone H3 is the loading control. D.TRF1 & TRF2 pull-down assay by telomere targeting CASID with the condition of DSP knockdown in HEK293 cells. E. Left, Dot blot of telomere DNA repeats in chromatin pull down by DSP with the condition of TRF1 or TRF2 knockdown. The dot-blot was performed at least three times & the represented one is shown. Right, the histogram shows the quantitated results of the dot blots. The triple asterisks (***) indicates a p-value less than 0.01. ns. indicates the insignificance difference. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31595153), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - A. TRF2 or DSP1 pull down by telomere targeting CASID with the condition of MST-312 treatment (left) or hTERT knockdown (right) in MDA-MB-231cells. B. Southern blot of telomere DNA from MDA-MB-231cells treated with MST-312 or with hTERT knockdown. C. Top, Dot blot of telomere DNA repeats from the chromatin pull down by DSP from the cells treated by MST-312 (left) or hTERT knockdown (right). Alu DNA repeats serve as negative control. Bottom, the histogram shows the quantitated results of the dot blots. The triple asterisks (***) indicates a p-value less than 0.01. D. Fluorescence image of DSP-EGFP transfected MDA-MB-231 cells with the conditions of MST-312 treatment or hTERT knockdown. E. Pull down assay of DSP by CASID targeting telomere in WI38 cells with or without ectopically expression of hTERT gene. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31595153), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Attenuation of bortezomib-induced shelterin protein dysregulation by hTERT over-expressionA. & B. Cells expressing ectopic hTERT were treated with bortezomib for 24 hours & mRNA levels of shelterin proteins then analyzed using qPCR. The levels of each target mRNA in bortezomib-treated cells were expressed as percentages of those in untreated cells. (A) HEL-hTERT cells & (B) BGC-823-hTERT cells. C. & D. Immunoblotting assessment of TRF1, TRF2 & POT1 protein expression in bortezomib-treated cells. Same sets of cells above were analyzed for TRF1, TRF2 & POT1 protein levels & shown was representative of three independent experiments. (C) HEL-hTERT cells & (D) BGC-823-hTERT cells.* & **: P < 0.05 & 0.01, respectively. BTZ, bortezomib. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.5752), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] -
Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] - RNaseH1 depletion leads to RPA activation at ALT telomeres.(a) HeLa & U2OS cells were transfected with the indicated siRNAs & 48 & 72 h later protein extracts were prepared. Western blot analysis was performed using antibodies against RNaseH1, RPA32 phosphorylated at Serine 33 (pSer33), total RPA32 & KAP1 (loading control). The asterisk indicates a cross-reacting band. Cells treated for 6 h with 5 mM hydroxyurea (HU) were used as controls for pSer33 activation. (b) SiRNA transfected cells were subjected to indirect immunofluorescence using antibodies against TRF2 (to visualize telomeres; red) & pSer33 (green). DNA was counterstained with DAPI (blue). Arrows point to examples of pSer33 foci co-localizing with TRF2 (TIFs). Scale bar, 9 μm. Cells treated for 6 h with 5 mM HU were used as controls for pSer33 activation. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25330849), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 interacts with lamin A/C.(a) Pull down of endogenous TRF2 in IMR90s. The top panel shows a blot probed for endogenous lamin A/C, & the bottom panel shows a blot with the same samples probed for TRF2. (b) Pull down of endogenously expressed lamin A/C in IMR90s. The top panel shows a blot probed for endogenous TRF2, & the bottom panel shows a blot with the same samples probed for endogenous lamin A/C. (c) Pull down of endogenous lamin A/C in IMR90s exogenously expressing GFP, GFP-TRF2, GFP-TRF deltaB deltaM or GFP-TRF1. The top panel shows a blot probed for GFP, & the bottom panel shows the same blot probed for lamin A/C. (d) Pull down of endogenous TRF2 in IMR90s exogenously expressing GFP, GFP-LMNA or progerin (GFP-LMNA delta50). The top panel shows a blot probed for GFP, & the bottom panel shows the same blot probed for TRF2. Asterisks denote IgG bands. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25399868), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Widespread dysregulation of shelterin protein expression in bortezomib-treated HEL & BGC-823 cellsA. & B. mRNA levels of shelterin factors TRF1, TRF2, TPP1, POT1, RAP1 & TIN2 in bortezomib-treated cells. Cells were treated with bortezomib for 24 hours & qPCR was used for quantitative assays. The levels of each target mRNA in bortezomib-treated cells were expressed as percentages of those in untreated cells. (A) HEL cells & (B) BGC-823 cells. C. & D. Immunoblotting assessment of TRF1, TRF2 & POT1 protein expression in bortezomib-treated cells. Same sets of cells above were analyzed for TRF1, TRF2 & POT1 protein levels & shown was representative of three independent experiments. (C) HEL cells & (D) BGC-823 cells. * & **: P < 0.05 & 0.01, respectively. BTZ, bortezomib. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.5752), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 interacts with lamin A/C.(a) Pull down of endogenous TRF2 in IMR90s. The top panel shows a blot probed for endogenous lamin A/C, & the bottom panel shows a blot with the same samples probed for TRF2. (b) Pull down of endogenously expressed lamin A/C in IMR90s. The top panel shows a blot probed for endogenous TRF2, & the bottom panel shows a blot with the same samples probed for endogenous lamin A/C. (c) Pull down of endogenous lamin A/C in IMR90s exogenously expressing GFP, GFP-TRF2, GFP-TRF deltaB deltaM or GFP-TRF1. The top panel shows a blot probed for GFP, & the bottom panel shows the same blot probed for lamin A/C. (d) Pull down of endogenous TRF2 in IMR90s exogenously expressing GFP, GFP-LMNA or progerin (GFP-LMNA delta50). The top panel shows a blot probed for GFP, & the bottom panel shows the same blot probed for TRF2. Asterisks denote IgG bands. Image collected & cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/25399868), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 transcriptionally regulates p21 expression through promoter occupancy. (A) Quantitative ChIP using TRF2 antibody gives enriched binding of TRF2 on p21 promoter in HT1080 & MDA-MB-231 cells, IgG was used as isotypic antibody control; normalized with 10% input (data represented as mean ± SEM, for three replicates). (B,C) TRF2 represses p21 promoter activity. In luciferase assay, si-RNA-mediated silencing or TRF2 over expression resulted in increased (B, *p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) or reduction (C, *p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) in p21 promoter activity in HT1080 & MDA-MB-231 cells, respectively; over expression of TRF2 devoid of DNA binding [deletion of basic (delB), myb (delM) & both basic/myb (delB-delM) domains] resulted in partial or complete rescue of p21 promoter activity in HT1080 & MDA-MB-231 cells (data represented as mean ± SEM, for three replicates) (C). (D,E) HT1080 cells over expressing TRF2 show reduced p21 protein expression, bar graph shows the densitometry analysis of three different immunoblot replicates (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates). (E) (Full images are shown in Supplementary Figure S5); TRF2 silencing results in increase in p21 protein, bar graph shows the densitometry analysis of three different immunoblots (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) (F); & mRNA expression in HT1080 cells (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates, GAPDH used as internal control for real-time PCR). Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-11177-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF2 transcriptionally regulates p21 expression through promoter occupancy. (A) Quantitative ChIP using TRF2 antibody gives enriched binding of TRF2 on p21 promoter in HT1080 & MDA-MB-231 cells, IgG was used as isotypic antibody control; normalized with 10% input (data represented as mean ± SEM, for three replicates). (B,C) TRF2 represses p21 promoter activity. In luciferase assay, si-RNA-mediated silencing or TRF2 over expression resulted in increased (B, *p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) or reduction (C, *p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) in p21 promoter activity in HT1080 & MDA-MB-231 cells, respectively; over expression of TRF2 devoid of DNA binding [deletion of basic (delB), myb (delM) & both basic/myb (delB-delM) domains] resulted in partial or complete rescue of p21 promoter activity in HT1080 & MDA-MB-231 cells (data represented as mean ± SEM, for three replicates) (C). (D,E) HT1080 cells over expressing TRF2 show reduced p21 protein expression, bar graph shows the densitometry analysis of three different immunoblot replicates (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates). (E) (Full images are shown in Supplementary Figure S5); TRF2 silencing results in increase in p21 protein, bar graph shows the densitometry analysis of three different immunoblots (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates) (F); & mRNA expression in HT1080 cells (*p value < 0.05, Student’s t-test; data represented as mean ± SEM of three replicates, GAPDH used as internal control for real-time PCR). Image collected & cropped by CiteAb from the following publication (https://www.nature.com/articles/s41598-017-11177-1), licensed under a CC-BY license. Not internally tested by Novus Biologicals.
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] -
Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - Attenuation of bortezomib-induced shelterin protein dysregulation by hTERT over-expressionA. & B. Cells expressing ectopic hTERT were treated with bortezomib for 24 hours & mRNA levels of shelterin proteins then analyzed using qPCR. The levels of each target mRNA in bortezomib-treated cells were expressed as percentages of those in untreated cells. (A) HEL-hTERT cells & (B) BGC-823-hTERT cells. C. & D. Immunoblotting assessment of TRF1, TRF2 & POT1 protein expression in bortezomib-treated cells. Same sets of cells above were analyzed for TRF1, TRF2 & POT1 protein levels & shown was representative of three independent experiments. (C) HEL-hTERT cells & (D) BGC-823-hTERT cells.* & **: P < 0.05 & 0.01, respectively. BTZ, bortezomib. Image collected & cropped by CiteAb from the following publication (https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.5752), licensed under a CC-BY license. Not internally tested by Novus Biologicals.

![Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Immunocytochemistry-Immunofluorescence-NB110-57130-img0011.jpg)
![Simple Western: TRF-2 AntibodyBSA Free [NB110-57130] Simple Western: TRF-2 AntibodyBSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Simple-Western-NB110-57130-img0010.jpg)
![Western Blot: TRF-2 AntibodyBSA Free [NB110-57130] Western Blot: TRF-2 AntibodyBSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Western-Blot-NB110-57130-img0008.jpg)
![Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130] Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Immunohistochemistry-Paraffin-NB110-57130-img0007.jpg)
![Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130] Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Immunohistochemistry-Paraffin-NB110-57130-img0013.jpg)
![Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Immunocytochemistry-Immunofluorescence-NB110-57130-img0016.jpg)
![Flow Cytometry: TRF-2 Antibody - BSA Free [NB110-57130] Flow Cytometry: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Flow-Cytometry-NB110-57130-img0017.jpg)
![Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Immunocytochemistry-Immunofluorescence-NB110-57130-img0015.jpg)
![Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130] Immunohistochemistry-Paraffin: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Immunohistochemistry-Paraffin-NB110-57130-img0012.jpg)
![Flow Cytometry: TRF-2 Antibody - BSA Free [NB110-57130] Flow Cytometry: TRF-2 Antibody - BSA Free [NB110-57130]](https://resources.bio-techne.com/images/products/TRF-2-Antibody-Flow-Cytometry-NB110-57130-img0014.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-210202423452420.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241528452.jpg)
![Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202415293320.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202415363710.jpg)
![Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202415371955.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202415284520.jpg)
![Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241538415.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202415392522.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624926.jpg)
![Chromatin Immunoprecipitation: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-3102024162491.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235527.jpg)
![Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235545.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624924.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624912.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235540.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241623553.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624919.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235510.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624913.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624911.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235549.jpg)
![Immunocytochemistry/ Immunofluorescence: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235532.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624921.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235531.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-3102024162490.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235546.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-31020241624927.jpg)
![Western Blot: TRF-2 Antibody - BSA Free [NB110-57130] - TRF-2 Antibody - BSA Free](https://resources.bio-techne.com/images/products/nb110-57130_rabbit-polyclonal-trf-2-antibody-310202416235529.jpg)