Recombinant SARS-CoV-2 Spike His-tag Biotin Protein, CF
R&D Systems, part of Bio-Techne | Catalog # BT10549

Key Product Details
Source
Accession #
Structure / Form
Conjugate
Applications
Product Specifications
Source
Val16-Lys1211 (Arg682Ser, Arg685Ser, Lys986Pro, Val987Pro), with a C-terminal 6-His tag
Purity
Endotoxin Level
N-terminal Sequence Analysis
Predicted Molecular Mass
SDS-PAGE
Activity
Measured by its binding ability in a functional ELISA with Recombinant Human ACE-2 Fc Chimera (Catalog # 10544-ZN).
Reviewed Applications
Read 1 review rated 5 using BT10549 in the following applications:
Scientific Data Images for Recombinant SARS-CoV-2 Spike His-tag Biotin Protein, CF
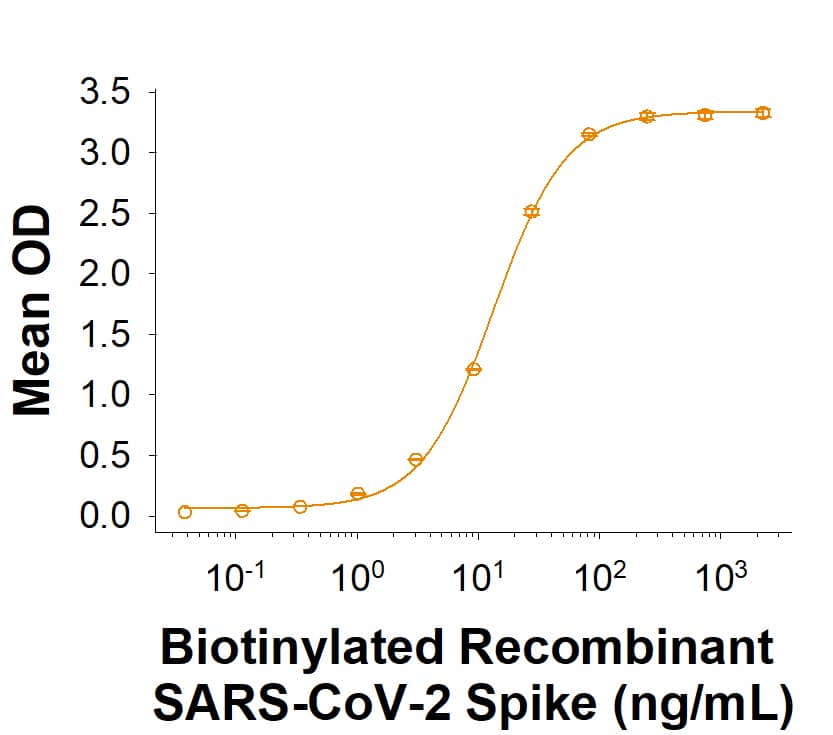
Recombinant SARS-CoV-2 Spike His-tag Biotinylated Protein Bioactivity.
Biotinylated Recombinant SARS-CoV-2 Spike His-tag (Catalog # BT10549) binds Recombinant Human ACE-2 Fc Chimera (10544-ZN) in a functional ELISA.Recombinant SARS-CoV-2 Spike His-tag Biotinylated Protein SDS-PAGE.
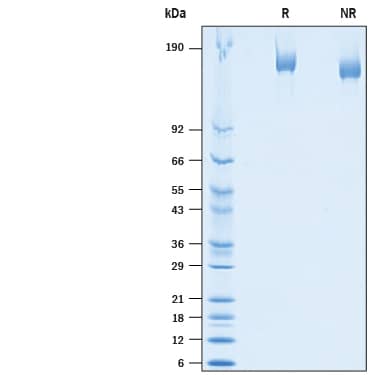
2 μg/lane of Biotinylated Recombinant SARS-CoV-2 Spike His-tag Protein (Catalog # BT10549) was resolved with SDS-PAGE under reducing (R) and non-reducing (NR) conditions and visualized by Coomassie® Blue staining, showing bands at 144-175 kDa.Detection of Biotinylated SARS-CoV-2 Spike Protein bound to ACE-2 expressing cells by flow cytometry.
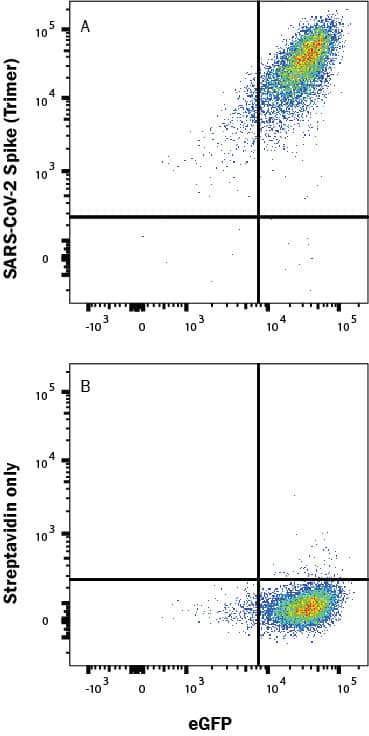
In a functional flow cytometry test, (A) Recombinant SARS-CoV-2 Spike His-tag Biotin Protein (Catalog # BT10549; 500 ng/ml) binds to HEK293 human embryonic kidney cell line transfected with recombinant human ACE-2 and EGFP. Ligand binding was detected by staining cells with APC-conjugated Streptavidin (F0050), which does not stain the cells in the absence of recombinant protein (B).Formulation, Preparation and Storage
BT10549
| Formulation | Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. |
| Reconstitution | Reconstitute at 500 μg/mL in PBS. |
| Shipping | The product is shipped at ambient temperature. Upon receipt, store it immediately at the temperature recommended below. |
| Stability & Storage | Use a manual defrost freezer and avoid repeated freeze-thaw cycles.
|
Background: Spike
SARS-CoV-2, which causes the global pandemic coronavirus disease 2019 (Covid-19), belongs to a family of viruses known as coronaviruses that are commonly comprised of four structural proteins: Spike protein (S), Envelope protein (E), Membrane protein (M), and Nucleocapsid protein (N) (1). SARS-CoV-2 Spike Protein (S Protein) is a glycoprotein that mediates membrane fusion and viral entry. The S protein is homotrimeric, with each ~180-kDa monomer consisting of two subunits, S1 and S2 (2). In SARS-CoV-2, as with most coronaviruses, proteolytic cleavage of the S protein into the S1 and S2 subunits is required for activation. The S1 subunit is focused on attachment of the protein to the host receptor while the S2 subunit is involved with cell fusion (3-5). The S protein of SARS-CoV-2 shares 75% and 29% amino acid (aa) sequence identity with the S protein of SARS-CoV-1 and MERS, respectively. The S Protein of the SARS-CoV-2 virus, like the SARS-CoV-1 counterpart, binds Angiotensin-Converting Enzyme 2 (ACE2), but with much higher affinity and faster binding kinetics through the receptor binding domain (RBD) located in the C-terminal region of S1 (6). Based on structural biology studies, the RBD can be oriented either in the up/standing or down/lying state with the up/standing state associated with higher pathogenicity (7). Polyclonal antibodies to the RBD of the SARS-CoV-2 protein have been shown to inhibit interaction with the ACE2 receptor, confirming RBD as an attractive target for vaccinations or antiviral therapy (8). It has been demonstrated that the S Protein can invade host cells through the CD147/EMMPRIN receptor and mediate membrane fusion (9, 10). A SARS-CoV-2 variant carrying the S protein aa change D614G has become the most prevalent form in the global pandemic and has been associated with greater infectivity and higher viral load (11, 12).
References
- Wu, F. et al. (2020) Nature 579:265.
- Tortorici, M.A. and D. Veesler (2019). Adv. Virus Res. 105:93.
- Bosch, B.J. et al. (2003). J. Virol. 77:8801.
- Belouzard, S. et al. (2009) Proc. Natl. Acad. Sci. 106:5871.
- Millet, J.K. and G.R. Whittaker (2015) Virus Res. 202:120.
- Ortega, J.T. et al. (2020) EXCLI J. 19:410.
- Yuan, Y. et al. (2017) Nat. Commun. 8:15092.
- Tai, W. et al. (2020) Cell. Mol. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Wang, X. et al. (2020) https://doi.org/10.1038/s41423-020-0424-9.
- Wang, K. et al. (2020) bioRxiv https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1.
- Korber, B. et al. (2020) Cell 182, 812.
- Zhang, L. et al. (2020) bioRxiv https://www.biorxiv.org/content/10.1101/2020.06.12.148726v1.
Long Name
Alternate Names
Entrez Gene IDs
Gene Symbol
UniProt
Additional Spike Products
Product Documents for Recombinant SARS-CoV-2 Spike His-tag Biotin Protein, CF
Product Specific Notices for Recombinant SARS-CoV-2 Spike His-tag Biotin Protein, CF
For research use only