By Emily Cartwright, PhD
How Can Psychological Stress Alter Your DNA?
Traumatic events, work demands, relationship conflicts, and health problems are all examples of psychological stressors that can result in physiological changes including increased cortisol and adrenaline levels. This change in gene expression manifests in physical symptoms such as increased heart rate, blood pressure, headaches, fatigue, muscle pain, and insomnia. In addition to changing gene expression in the short term, psychological stress can impart lasting changes to your DNA by modifying the epigenome.
The prefix “epi” means “above”, referring to changes “above” or “on” the DNA structure itself, rather than within the DNA sequence. Studies have shown that social stress can cause changes to the structure of DNA, with changes in DNA methylation and patterns of histone methylation and acetylation. These changes persist during cell division and are passed on to the daughter cells. Collectively, we refer to these changes to the DNA structure as epigenetics. Much of our understanding of how stress and epigenetics relates to mental health issues like depression and anxiety, comes from studies in mice and rat brains, with some corroborating evidence in post-mortem human brain tissue.
A Link Between DNA Methylation and Depression?
In general, DNA methylation is associated with a “closed” gene, or repression of gene expression. Changes in DNA methylation have been associated with chronic social defeat in mice, a stress model which induces depression-like symptoms. Research has found that chronic social defeat increases expression of DNMT3A in the nucleus accumbens (NAc) and reducing expression with an inhibitor, like RG108, imparts antidepressant-like effects.1
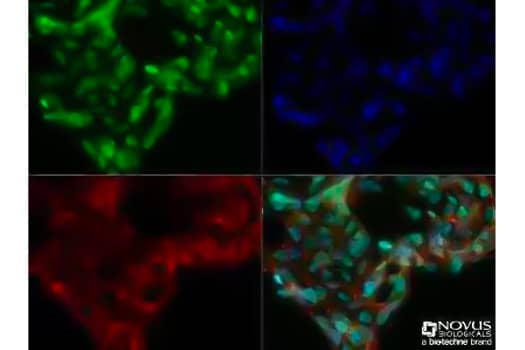
Immunocytochemistry/Immunofluorescence: Ntera2 cells were fixed for 10 minutes using 10% formalin and then permeabilized for 5 minutes using 1X TBS + 0.5% Triton-X100. The cells were incubated with Mouse Anti-DNMT3A (68B1446) (Catalog # NB120-13888) at a 1:200 dilution overnight at 4C and detected with an anti-mouse Dylight 488 (Green) at a 1:500 dilution. Actin was detected with Phalloidin 568 (Red) at a 1:200 dilution. Nuclei were counterstained with DAPI (Blue). Cells were imaged using a 40X objective. This antibody has been validated by Biological Strategies.
DNMT3A expression is most often associated with transcriptional repression, so increased expression of the gene would most likely lead to decreased expression of other genes. Chronic social defeat is also observed with an associated with decrease in expression of NR3C1, BDNF, and SLC6A4. NR3C1 encodes the glucocorticoid receptor (GR), and decreased expression at this locus is associated with reduced GR expression and reduced ability to provide negative feedback to the hypothalamus and pituitary gland, leading to hyperactivity of the hypothalamic-pituitary-adrenal (HPA) axis.2 Increased methylation of SLC6A4 can increase amygdala reactivity and risk of depression by decreasing expression of serotonin transporter.2
Modulating Depressive Symptoms with Histone Modifications
The earliest report of an epigenetic component to depression came from the observation that inhibitors of the family of enzymes responsible for histone deacetylation (HDACs) had antidepressant-like effects in mice.3,4 Histone acetylation is carried out by histone acetyltransferases (HATs). These enzymes deposit acyl groups onto lysine residues present on histone tails, allowing for the chromatin structure to open. HATs are transcriptional facilitators, while HDACs that remove acyl groups, are transcriptional repressors.
Selected HDAC inhibitors
| Inhibitor | Target | Bio-Techne Catalog # |
|---|---|---|
| SAHA | Class I and Class II HDAC | 4652 |
| Sodium butyrate | General HDAC | 3850 |
| MS 275 | Class I HDAC | 6208 |
Unlike DNA methylation, histone methylation can either activate or repress transcription depending on the amino acid (usually lysine (K) or arginine (R)) and the number of methyl groups. For example, trimethylation of histone H3 at lysine 4 (H3K4me3) is an active mark of transcription, whereas dimethylation of histone H3 at lysine 9 (H3K9me2), is transcriptionally silencing. Importantly for epigenetics and depression, H3K9me2 is an repressive mark within the NAc. Chronic social defeat in mice downregulates G9a, the methyltransferase responsible for catalyzing H3K9me2. More specifically, reduction of H3K9me2 at specific loci like Ras, increases ERK signaling and depressive symptoms. Increase in another histone mark, H3K27me2, upstream of Rac1 has been associated with depressive behavior in both mice and humans.
While exposure to chronic stress and stress early in life can change the epigenetics of certain important genes, most, if not all, epigenetic changes are reversible. The more we understand this relationship, the better positioned we can be to develop novel treatments and improve mental health for all.
![Chromatin Immunoprecipitation (ChIP) analysis of Rabbit Anti-Histone H3 [Dimethyl Lys9] used to IP chromatin from Hela cells and compared to a no antibody control. Target gene immunoprecipitated DNA gene expression was measured using qRT-PCR and SYBR gree](https://resources.bio-techne.com/bio-techne-assets/images/blog/epigenetics-of-depression-fig-3.png)
Chromatin Immunoprecipitation (ChIP) analysis of Rabbit Anti-Histone H3 [Dimethyl Lys9] Polyclonal Antibody (Catalog # NB21-1072) used to IP chromatin from fixed Hela cells alongside a no antibody (No Ab) control. Target gene immunoprecipitated DNA was measured by qRT-PCR and SYBR green dye, then normalized to the non-precipitated input chromatin (input=1).

Emily Cartwright, PhD
Product Marketing Specialist, Antibodies, at Novus Biologicals LLC
-
LaPlant, Q. et al. (2010) Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens Nat Neurosci 13:1137-1143.
-
Cunliffe, V.T. (2016) The epigenetic impacts of social stress: how does social adversity become biologically embedded? Epigenomics 8:1653-1669.
-
Tsankova, N.M. et al. (2006) Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action Nat Neurosci 9:519-525.
-
Schroeder, F.A. et al. (2007) Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse Biol Psychiatry 62:55-64.