Protein Degraders
Targeted Protein Degradation requires the use of Protein Degraders to achieve potent and selective knock-down of target proteins at the post-translational level. A growing number of different modalities have emerged that can bring about targeted protein knockdown, with small molecule bifunctional Degraders, such as PROTAC® molecules, SNIPERs, PHOTACs, Degronimids and uSMITEs™, at the forefront of the field.
Bio-Techne provides a wide range of Protein Degraders and negative controls to help you investigate your target of interest.
What are Small Molecule Protein Degraders?
Protein Degraders are bifunctional compounds comprising 3 components: an E3 ubiquitin ligase ligand, a linker, and a ligand for a target protein of interest. They induce the formation of a ternary complex by simultaneously binding to both an E3 ligase and target protein. Ternary complex formation effectively ‘hijacks’ the E3 ligase to polyubiquitinate a target of interest, inducing subsequent degradation by the 26S proteasome.
Degraders are attractive tools for use in basic research to induce selective protein knockdown in a reversible and tuneable manner, without the requirement for genetic modification to cells. Degraders also have therapeutic potential as an approach to target the ‘undruggable’ proteome and overcome common resistance mechanisms to current therapies.
The PROTAC® concept was first described in 2001. The first generation of Degraders synthesized were peptide-based and were not cell-permeable, however they demonstrated the principle that it is possible to direct an E3 ligase to degrade a protein of interest. The first small molecule protein Degraders were also reported by Craig Crews’ group in 2008. Since then, a wide range of degraders have been synthesized, recruiting different E3 ligase enzymes, including Cereblon, von Hippel Lindau (VHL) and inhibitor of apoptosis (IAP), and targeting a variety of protein targets, such as enzymes, receptors, bromodomains and transcription factors.
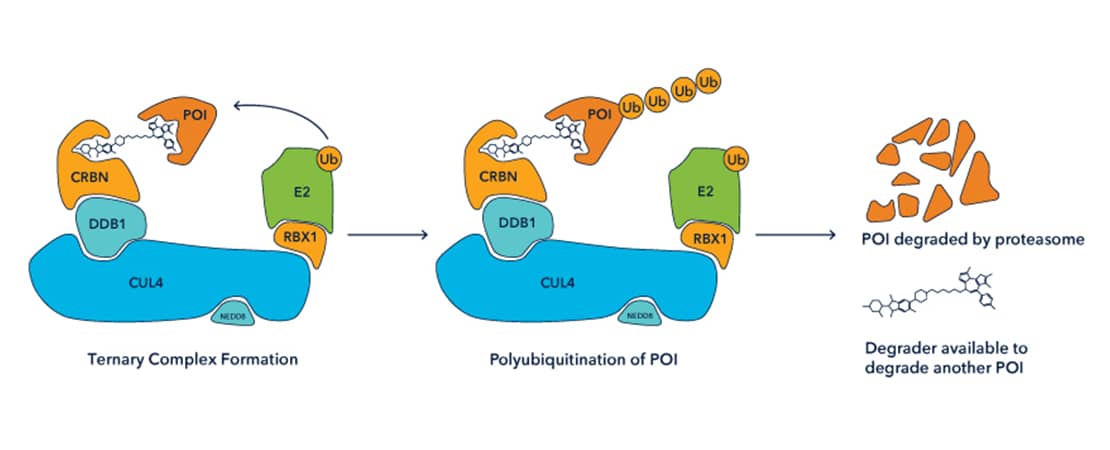

Figure 2: Wes data showing knockdown of both CDK9 isoforms after THAL SNS 032 (Cat. No. 6532) treatment (200 or 500 nM) of MOLT-4 cells. Protein quantification (relative to DMSO-only control) is shown beneath the corresponding lane.
Experiments were performed by the Simple Western™ applications science team. Simple Western fully automated western blotting systems provide quantitative data, which allows for an accurate determination of DC50 and Dmax values for Protein Degraders.
Advantages of Protein Degraders
Protein Degraders offer a convenient alternative to methods such as RNAi and CRISPR gene editing for achieving protein knockdown. Some advantages are summarized below:
- Degraders are easy and quick to use. They can be applied directly to cells with no requirement for expression vectors or transfection.
- The level of protein degradation can be easily tuned by altering the dose of Degrader used.
- Degraders can be used in a broad range of different cell lines, as they do not require cells that are easily transfectable.
- The effects of Degraders can be reversed on washout, so the duration of effect can be readily controlled.
- Degraders can elicit pronounced and sustained effects at sub-stoichiometric concentrations (this is a result of their catalytic mode of action). Degraders can offer enhanced selectivity compared to small molecule inhibitors. For example, the PROTAC® molecule AT 1 (Cat. No. 6356), is a selective degrader of BRD4, whereas the target protein ligand component of AT 1, (+)- JQ1 (Cat. No. 4499), also inhibits closely-related BET bromodomain family proteins.
Using Protein Degraders
The activity of Degraders is represented by the DC50 and Dmax values. The DC50 is a measure of potency and is the concentration at which 50% of the target protein is degraded; it is equivalent to the IC50 for an inhibitor. The Dmax is the maximum level of degradation achievable. These values are useful guides to determine the appropriate concentration of Degrader to use in your experiment. The timescale to achieve maximum degradation varies significantly between different Degraders and should also be taken into consideration when planning your experiment.
Effective knockdown of target proteins can be achieved with very low Degrader concentrations. In some cases, Degraders exhibit the “hook effect” at higher doses. This effect refers to the reduced degradation efficiency resulting from the increased formation of binary complexes at higher Degrader concentrations, which compete with formation of ternary complexes.

Figure 3: The so-called “hook effect” is demonstrated in this western blot, which shows Degrader-mediated polyubiquitination of recombinant BRD4. CRBN E3 ligase complex (Cat. No. E3-650) was used to investigate Degrader-induced in vitro polyubiquitination of recombinant FLAG-tagged BRD4 (Cat. No. SP-600). Results were analyzed using an anti-FLAG Western Blot. The degree of substrate ubiquitination varies with the concentration of Degrader used in the reaction.
Molecular Glues
A Molecular Glue is a small molecule protein dimerizers that brings about targeted protein degradation via a similar mechanism to PROTAC® Degraders. However, molecular glues are not chimeric molecules. Molecular glues act by forming a ternary complex with a target protein and an E3 ligase leading to ubiquitination of the protein and its subsequent destruction by the proteasome. The interaction of the molecular glue with the E3 ligase may result in allosteric modification of the E3 ligase binding site, such that it is unable to bind its native substrate, enabling ternary complex formation with a protein of interest (or neosubstrate). Like other small molecule Protein Degraders, molecular glues act catalytically.
Background Information
- LYTAC ( (LYsosome TArgeting Chimera molecules): LYTACs direct an extracellular protein of interest to the lysosome for degradation and were invented at the University of Wisconsin
- PROTAC® (PROteolysis TArgeting Chimeric molecules): this term was coined in a paper from Craig Crews’ lab (Yale University) in a 2001 paper and is now widely used to refer to protein degrader molecules.
- SNIPERs (Specific and Nongenetic Inhibitor of apoptosis protein (IAP)-dependent Protein ERasers): SNIPERs specifically recruit the E3 ligase IAP and were developed at the University of Tokyo.
- PHOTAC (PHOtochemically TArgeting Chimeras): these are photoswitchable degraders that can be activated by different wavelengths of light and were originated at New York University.
- uSMITE™ (Ubiquitin-mediated, Small Molecule-Induced Target Elimination technology): a type of Degrader developed by Cullgen
-
Itoh, Y et al. (2010) Protein knockdown using methyl bestatin-ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J. Am. Chem. Soc . 132:5820. PMID: 20369832.
-
Reynders, M et al. (2020) PHOTACs enable optical control of protein degradation. Sci. Adv. PMID: 32128406.
-
Rodriguez-Gonzalez, A et al. (2008) Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene . PMID: 18794799.
-
Sakamoto, K et al. (2001) Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA PMID: 11438690.
-
Schneekloth, A et al. (2008) Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg. Med. Chem. Lett. PMID: 18752944.
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.