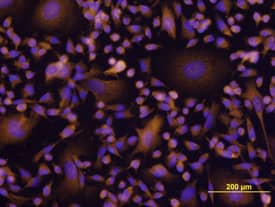
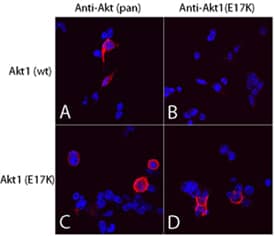
Akt (pan) in 293T Human Cell Line.
Akt pan specific (panels A and C) and Akt1 (E17K Mutation) (panels B and D) were detected in immersion fixed 293T human embryonic kidney cell line transfected with wild type (panels A and B) or E17K mutated (panels C and D) Akt1 using Mouse Anti-Human/Mouse/Rat Akt Pan Specific Monoclonal Antibody (Catalog # MAB2055) and Mouse Anti-Human Akt1 (E17K Mutation) Monoclonal Antibody (Catalog #
MAB6815). Both antibodies were used at 10 µg/mL for 3 hours at room temperature. Cells were stained using the NorthernLights™ 557-conjugated Anti-Mouse IgG Secondary Antibody (red; Catalog #
NL007) and counterstained with DAPI (blue). Specific staining was localized to plasma membranes and cytoplasm. View our protocol for Fluorescent ICC Staining of Cells on Coverslips.
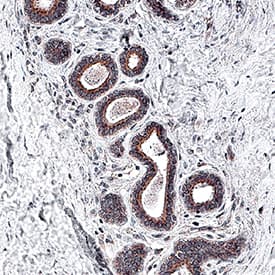
Akt Pan Specific in Human Breast Cancer Tissue.
Akt was detected in immersion fixed paraffin-embedded sections of human breast cancer tissue using Mouse Anti-Human/Mouse/Rat Akt Pan Specific Monoclonal Antibody (Catalog # MAB2055) at 5 µg/mL for 1 hour at room temperature followed by incubation with the Anti-Mouse IgG VisUCyte™ HRP Polymer Antibody (
VC001). Before incubation with the primary antibody, tissue was subjected to heat-induced epitope retrieval using Antigen Retrieval Reagent-Basic (
CTS013). Tissue was stained using DAB (brown) and counterstained with hematoxylin (blue). Specific staining was localized to cytoplasm in epithelial cells. Staining was performed using our IHC Staining with VisUCyte HRP Polymer Detection Reagents.
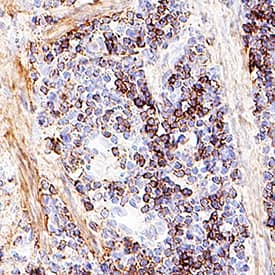
Akt Pan Specific in Mouse Spleen Tissue.
Akt was detected in immersion fixed paraffin-embedded sections of mouse spleen tissue using Mouse Anti-Human/Mouse/Rat Akt Pan Specific Monoclonal Antibody (Catalog # MAB2055) at 5 µg/mL for 1 hour at room temperature followed by incubation with the Anti-Mouse IgG VisUCyte™ HRP Polymer Antibody (
VC001). Before incubation with the primary antibody, tissue was subjected to heat-induced epitope retrieval using Antigen Retrieval Reagent-Basic (
CTS013). Tissue was stained using DAB (brown) and counterstained with hematoxylin (blue). Specific staining was localized to cytoplasm in lymphocytes. Staining was performed using our IHC Staining with VisUCyte HRP Polymer Detection Reagents.
Detection of Akt in MCF-7 Human Cell Line by Flow Cytometry.
MCF-7 human breast cancer cell line was stained with Mouse Anti-Human/Mouse/Rat Akt Monoclonal Antibody (Catalog # MAB2055, filled histogram) or isotype control antibody (Catalog #
MAB0041, open histogram), followed by Phycoerythrin-conjugated Anti-Mouse IgG F(ab')
2Secondary Antibody (Catalog #
F0102B). To facilitate intracellular staining, cells were fixed with paraformaldehyde and permeabilized with saponin.
Detection of Human Akt by Simple WesternTM.
Simple Western lane view shows lysates of MCF‑7 human breast cancer cell line, loaded at 0.2 mg/mL. A specific band was detected for Akt at approximately 62 kDa (as indicated) using 2 µg/mL of Mouse Anti-Human/Mouse/Rat Akt Pan Specific Monoclonal Antibody (Catalog # MAB2055). This experiment was conducted under reducing conditions and using the 12-230 kDa separation system.
Detection of Human AKT by Western Blot
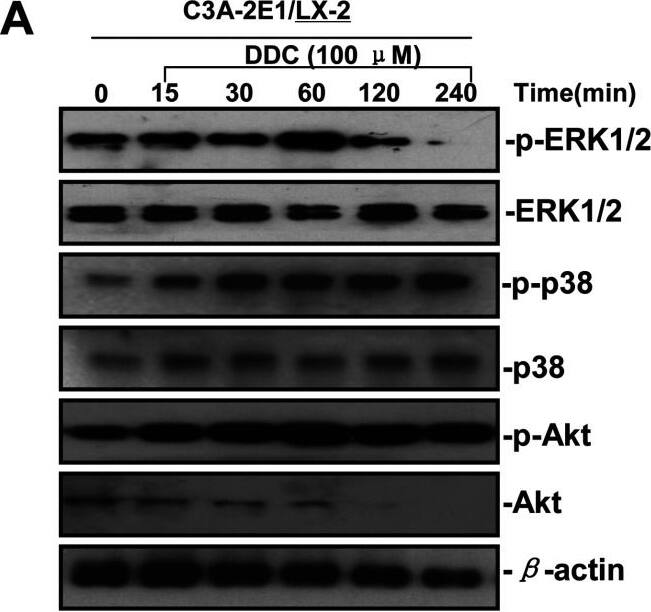
DDC up-regulates MMP-1 through ERK1/2 and Akt activation(A) Co-cultures were treated with 100 μM DDC for the indicated time. Phosphorylation of ERK1/2, p38 and Akt were determined by Western blotting analysis. The corresponding non-phosphorylated ERK1/2, p38, Akt and beta-actin were used for protein loading control. (B) Co-cultures were treated with or without 100 μM DDC for 24 h in the presence or absence of ERK1/2 inhibitor (U0126, 10 μM). MMP-1 protein levels were analysed by Western blotting. (C) Co-cultures were treated with or without 100 μM DDC for 24 h in the presence or absence of p38 inhibitor (SB203580, 10 μM). MMP-1 protein levels were analysed by Western blotting. (D) Co-cultures were treated with or without 100 μM DDC for 24 h in the presence or absence of Akt inhibitor (T3830, 50 μM). MMP-1 protein levels were analysed by Western blotting. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23577625), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human AKT by Western Blot
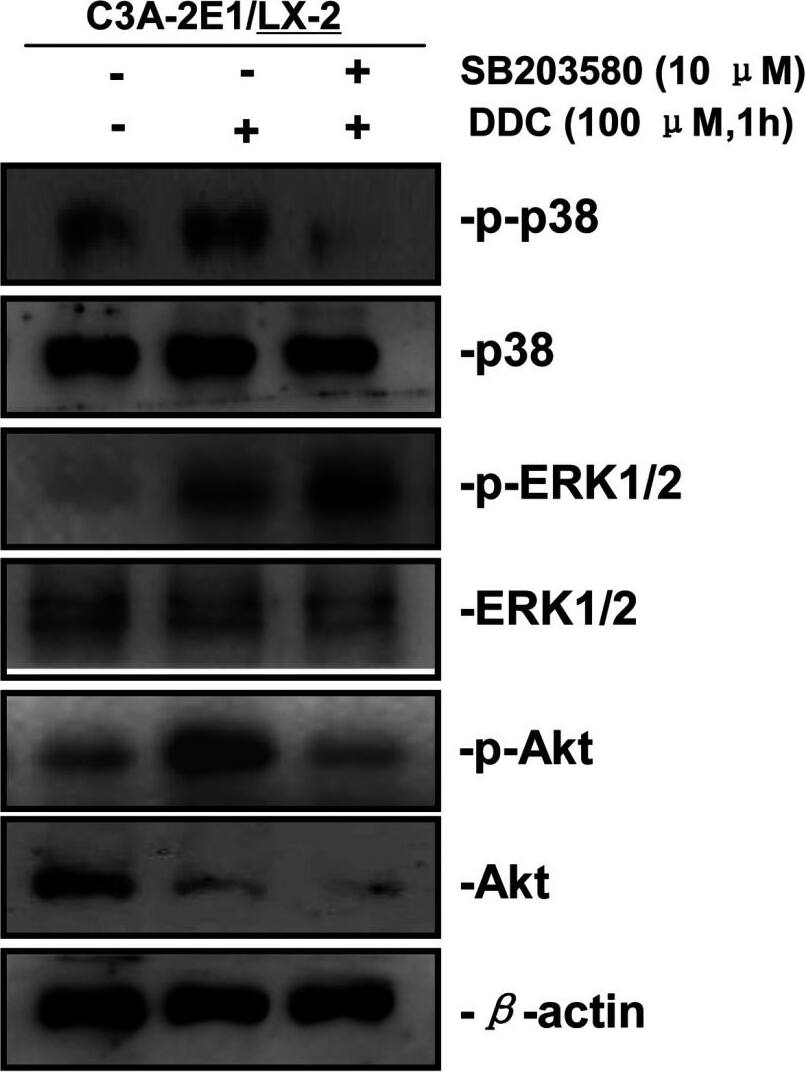
p38 inhibitor SB203580 improves the up-regulation of MMP-1 by DDC through stimulating ERK1/2Co-cultures were treated with or without 100 μM DDC for 1 h in the presence or absence of p38 inhibitor (SB203580, 10 μM). Phosphorylation of ERK1/2, p38 and Akt in LX-2 cells of co-cultures were determined by Western blotting analysis. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23577625), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse AKT by Western Blot
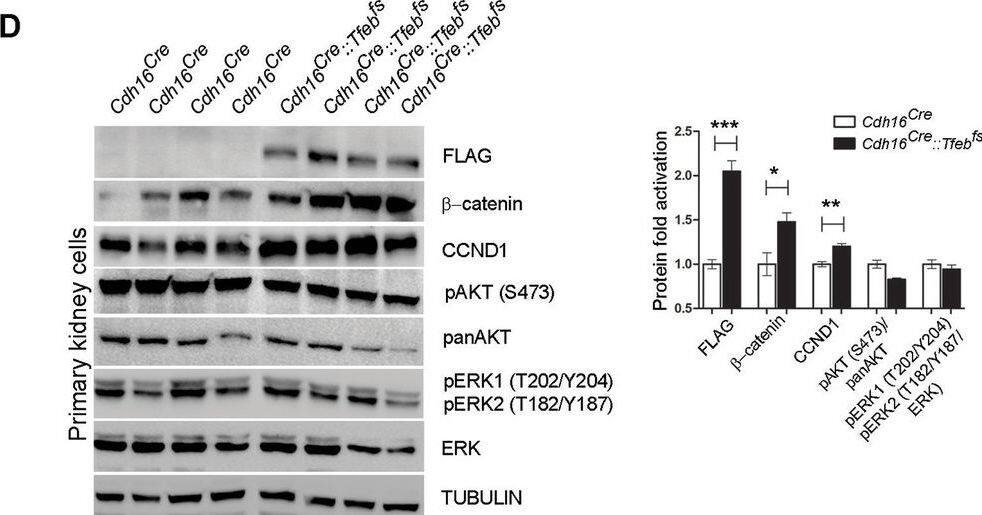
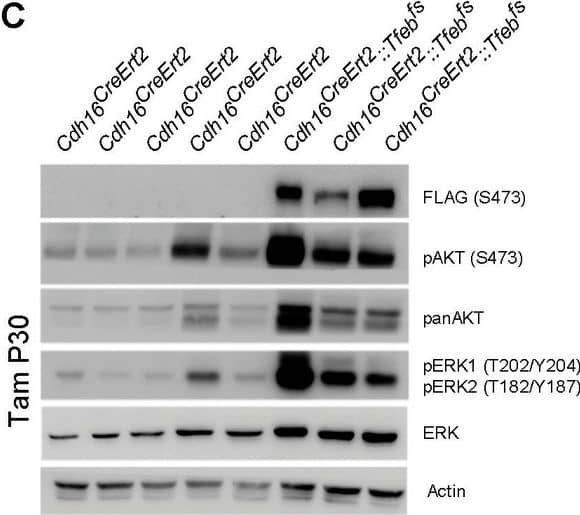
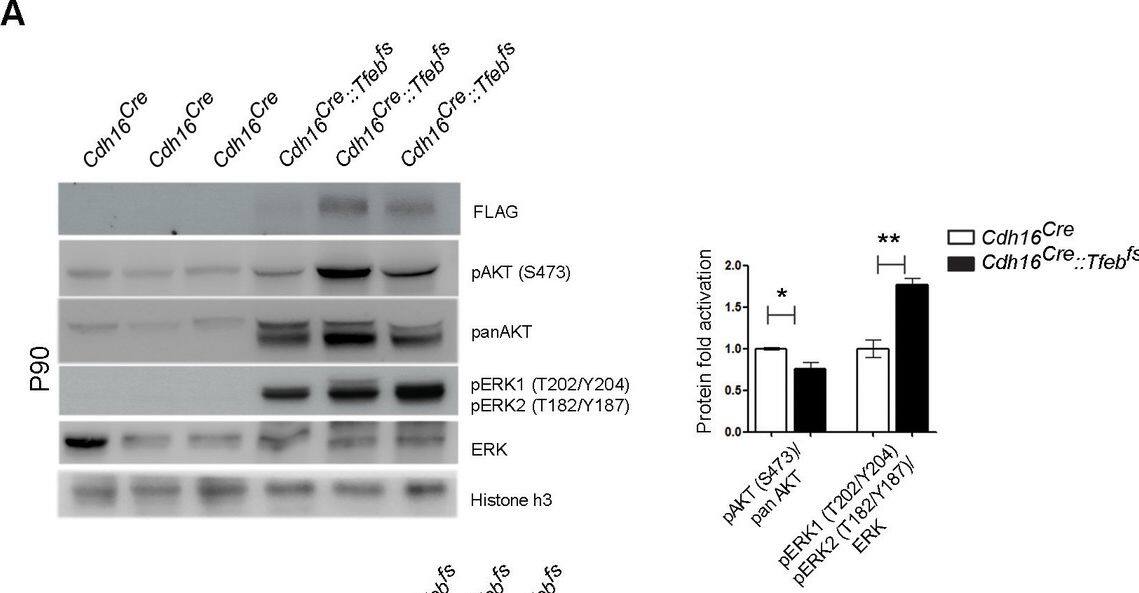
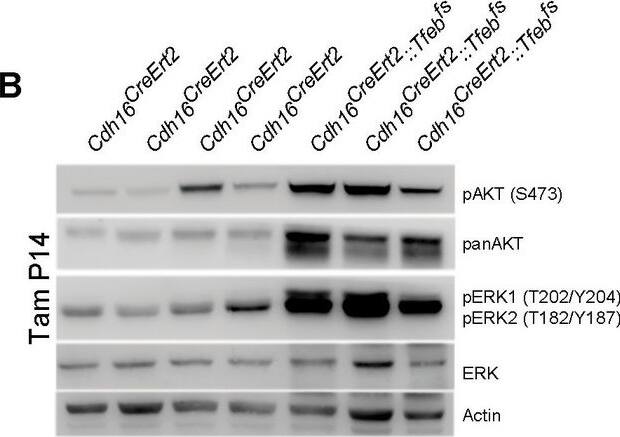
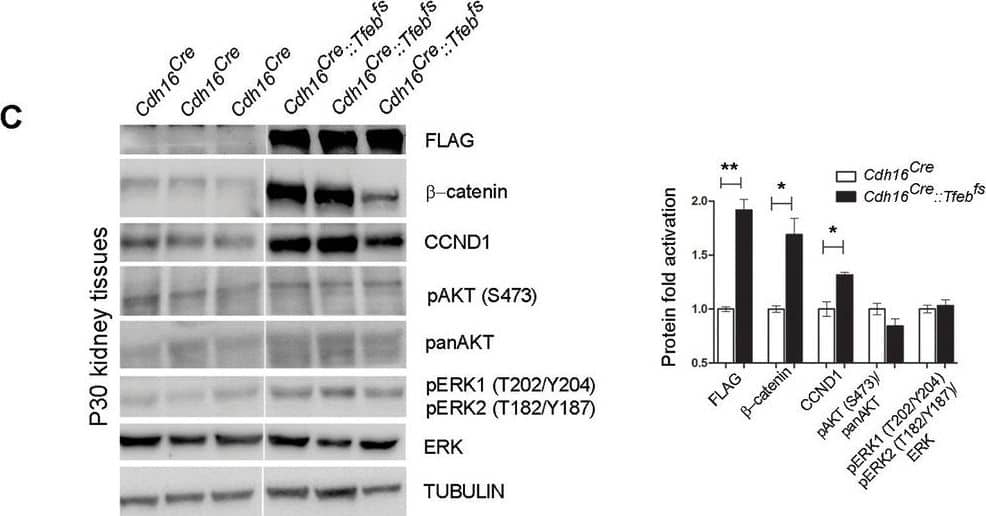
Activation of ErbB and WNT signaling pathways in kidneys from Cdh16Cre::Tfebfs mice.Transcriptional and biochemical analyses were performed on Cdh16Cre and Cdh16Cre::Tfebfs mice. (A,B) Tables show the relative increase of genes related to the ErbB (A) and WNT (B) pathways in the microarray analyses performed on kidneys from P0 Cdh16Cre::Tfebfs mice. Graphs show real-time PCR validations performed on kidneys from Cdh16Cre::Tfebfs mice at different stages (P0, P12, P30). Data are shown as the average (± SEM) of at least three Cdh16Cre::Tfebfs mice normalized versus wild-type mice. (C,D) Immunoblot analyses performed on (C) P30 kidney tissues and (D) primary kidney cells isolated from Cdh16Cre::Tfebfs mice to evaluate ErbB and WNT activation status. Each replicate is a distinct biological sample. ErbB signaling was assessed by looking at phosphoAKT (Ser473) to total AKT ratio, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK ratio; WNT signaling was assessed by quantifying beta-catenin and CCND1 (Cyclin D1) protein levels. Graphs represent the densitometry quantification of Western blot bands. Values are normalized to actin when not specified and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided, Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.00710.7554/eLife.17047.008Figure 3—source data 1.Complete list of 294 genes (represented by 361 probesets) significantly induced (FDR≤0.05) in the KSP_P0 microarray dataset (GSE62977).The genes are ranked by decreasing signed ratio (KSP_P0/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.00810.7554/eLife.17047.009Figure 3—source data 2.Complete list of 628 genes (represented by 729 probesets) significantly induced (FDR≤0.05) in the KSP_P14 microarray dataset (GSE63376).The genes are ranked by decreasing signed ratio (KSP_P14/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.009Complete list of 294 genes (represented by 361 probesets) significantly induced (FDR≤0.05) in the KSP_P0 microarray dataset (GSE62977).The genes are ranked by decreasing signed ratio (KSP_P0/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.008Complete list of 628 genes (represented by 729 probesets) significantly induced (FDR≤0.05) in the KSP_P14 microarray dataset (GSE63376).The genes are ranked by decreasing signed ratio (KSP_P14/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.009ErbB and WNT transcriptional profiles in Cdh16CreErt2::Tfebfs mice.Transcriptional analyses performed on Cdh16CreErt2::Tfebfs mice. (A,B) mRNA levels of previously validated genes belonging to the WNT (left graphs) and ErbB (right graphs) signaling pathways assessed in P90 Cdh16CreErt2::Tfebfs mice induced at (A) P14 and at (B) P30 with tamoxifen respectively. Data are shown as the average (± SEM) of at least Cdh16CreErt2::Tfebfs mice and values are normalized to the wild-type line. (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.010Biochemical analysis of ErbB signaling.Immunoblot analysis performed on P90 kidneys from Cdh16Cre::Tfebfs mice (A) and P90 Cdh16CreErt2::Tfebfs animals induced with tamoxifen at P14 (B) and at P30 (C), respectively. Each replicate is a different biological sample. ErbB was analyzed by quantifying phosphoAKT (Ser473) to total AKT, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK; graphs are the densitometry quantifications of Western blot bands normalized to wild-type line and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.011 Image collected and cropped by CiteAb from the following publication (https://elifesciences.org/articles/17047), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human AKT by Western Blot
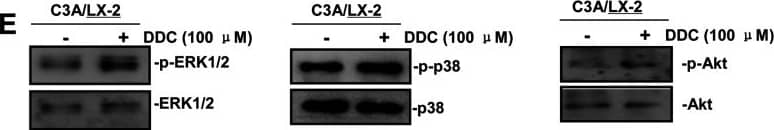
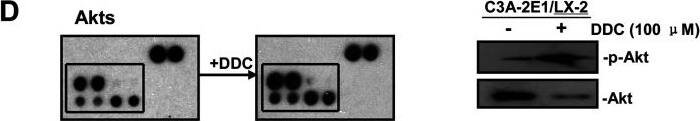
Phospho-proteome profiling results of LX-2 cells co-cultured with C3A–CYP2E1 cells with or without DDC treatment and detection of the effect of DDC on intracellular kinases in LX-2 cells of co-cultures200 μg of total cell lysates from LX-2 cells co-cultured with C3A-2E1 cells with or without 100 μM DDC for 1 h were incubated with membranes of the human phospho-MAPK Array Kit according to the manufacturer's instructions. Phospho MAPK Array data were developed on X-ray films following exposure to chemiluminescent reagents. 20 μg aliquots of total cell lysates from LX-2 cells were subjected to Western blotting analysis. (A) Template showing the location of MAPK antibodies spotted onto the human phospho-MAPK Array Kit. (B) The activation status of ERK1/2 in LX-2 cells co-cultured with C3A-2E1 cells after DDC treatment. (C) The activation status of p38 in LX-2 cells co-cultured with C3A-2E1 cells after DDC treatment. (D) The activation status of Akt in LX-2 cells co-cultured with C3A-2E1 cells after DDC treatment. (E) The activation status of ERK1/2, p38 and Akt in LX-2 cells co-cultured with C3A cells after DDC treatment. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23577625), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human AKT by Western Blot
Phospho-proteome profiling results of LX-2 cells co-cultured with C3A–CYP2E1 cells with or without DDC treatment and detection of the effect of DDC on intracellular kinases in LX-2 cells of co-cultures200 μg of total cell lysates from LX-2 cells co-cultured with C3A-2E1 cells with or without 100 μM DDC for 1 h were incubated with membranes of the human phospho-MAPK Array Kit according to the manufacturer's instructions. Phospho MAPK Array data were developed on X-ray films following exposure to chemiluminescent reagents. 20 μg aliquots of total cell lysates from LX-2 cells were subjected to Western blotting analysis. (A) Template showing the location of MAPK antibodies spotted onto the human phospho-MAPK Array Kit. (B) The activation status of ERK1/2 in LX-2 cells co-cultured with C3A-2E1 cells after DDC treatment. (C) The activation status of p38 in LX-2 cells co-cultured with C3A-2E1 cells after DDC treatment. (D) The activation status of Akt in LX-2 cells co-cultured with C3A-2E1 cells after DDC treatment. (E) The activation status of ERK1/2, p38 and Akt in LX-2 cells co-cultured with C3A cells after DDC treatment. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/23577625), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse AKT by Western Blot
Biochemical analysis of ErbB signaling.Immunoblot analysis performed on P90 kidneys from Cdh16Cre::Tfebfs mice (A) and P90 Cdh16CreErt2::Tfebfs animals induced with tamoxifen at P14 (B) and at P30 (C), respectively. Each replicate is a different biological sample. ErbB was analyzed by quantifying phosphoAKT (Ser473) to total AKT, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK; graphs are the densitometry quantifications of Western blot bands normalized to wild-type line and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.011 Image collected and cropped by CiteAb from the following publication (https://elifesciences.org/articles/17047), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse AKT by Western Blot
Biochemical analysis of ErbB signaling.Immunoblot analysis performed on P90 kidneys from Cdh16Cre::Tfebfs mice (A) and P90 Cdh16CreErt2::Tfebfs animals induced with tamoxifen at P14 (B) and at P30 (C), respectively. Each replicate is a different biological sample. ErbB was analyzed by quantifying phosphoAKT (Ser473) to total AKT, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK; graphs are the densitometry quantifications of Western blot bands normalized to wild-type line and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.011 Image collected and cropped by CiteAb from the following publication (https://elifesciences.org/articles/17047), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse AKT by Western Blot
Biochemical analysis of ErbB signaling.Immunoblot analysis performed on P90 kidneys from Cdh16Cre::Tfebfs mice (A) and P90 Cdh16CreErt2::Tfebfs animals induced with tamoxifen at P14 (B) and at P30 (C), respectively. Each replicate is a different biological sample. ErbB was analyzed by quantifying phosphoAKT (Ser473) to total AKT, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK; graphs are the densitometry quantifications of Western blot bands normalized to wild-type line and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.011 Image collected and cropped by CiteAb from the following publication (https://elifesciences.org/articles/17047), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Mouse AKT by Western Blot
Activation of ErbB and WNT signaling pathways in kidneys from Cdh16Cre::Tfebfs mice.Transcriptional and biochemical analyses were performed on Cdh16Cre and Cdh16Cre::Tfebfs mice. (A,B) Tables show the relative increase of genes related to the ErbB (A) and WNT (B) pathways in the microarray analyses performed on kidneys from P0 Cdh16Cre::Tfebfs mice. Graphs show real-time PCR validations performed on kidneys from Cdh16Cre::Tfebfs mice at different stages (P0, P12, P30). Data are shown as the average (± SEM) of at least three Cdh16Cre::Tfebfs mice normalized versus wild-type mice. (C,D) Immunoblot analyses performed on (C) P30 kidney tissues and (D) primary kidney cells isolated from Cdh16Cre::Tfebfs mice to evaluate ErbB and WNT activation status. Each replicate is a distinct biological sample. ErbB signaling was assessed by looking at phosphoAKT (Ser473) to total AKT ratio, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK ratio; WNT signaling was assessed by quantifying beta-catenin and CCND1 (Cyclin D1) protein levels. Graphs represent the densitometry quantification of Western blot bands. Values are normalized to actin when not specified and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided, Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.00710.7554/eLife.17047.008Figure 3—source data 1.Complete list of 294 genes (represented by 361 probesets) significantly induced (FDR≤0.05) in the KSP_P0 microarray dataset (GSE62977).The genes are ranked by decreasing signed ratio (KSP_P0/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.00810.7554/eLife.17047.009Figure 3—source data 2.Complete list of 628 genes (represented by 729 probesets) significantly induced (FDR≤0.05) in the KSP_P14 microarray dataset (GSE63376).The genes are ranked by decreasing signed ratio (KSP_P14/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.009Complete list of 294 genes (represented by 361 probesets) significantly induced (FDR≤0.05) in the KSP_P0 microarray dataset (GSE62977).The genes are ranked by decreasing signed ratio (KSP_P0/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.008Complete list of 628 genes (represented by 729 probesets) significantly induced (FDR≤0.05) in the KSP_P14 microarray dataset (GSE63376).The genes are ranked by decreasing signed ratio (KSP_P14/CTL).DOI:https://dx.doi.org/10.7554/eLife.17047.009ErbB and WNT transcriptional profiles in Cdh16CreErt2::Tfebfs mice.Transcriptional analyses performed on Cdh16CreErt2::Tfebfs mice. (A,B) mRNA levels of previously validated genes belonging to the WNT (left graphs) and ErbB (right graphs) signaling pathways assessed in P90 Cdh16CreErt2::Tfebfs mice induced at (A) P14 and at (B) P30 with tamoxifen respectively. Data are shown as the average (± SEM) of at least Cdh16CreErt2::Tfebfs mice and values are normalized to the wild-type line. (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.010Biochemical analysis of ErbB signaling.Immunoblot analysis performed on P90 kidneys from Cdh16Cre::Tfebfs mice (A) and P90 Cdh16CreErt2::Tfebfs animals induced with tamoxifen at P14 (B) and at P30 (C), respectively. Each replicate is a different biological sample. ErbB was analyzed by quantifying phosphoAKT (Ser473) to total AKT, and phosphoERK1 (T202/Y204)/ERK2(T185/Y187) to total ERK; graphs are the densitometry quantifications of Western blot bands normalized to wild-type line and are shown as an average (± SEM) (*p<0.05, **p<0.01, ***p<0.001, two-sided Student’s t test).DOI:https://dx.doi.org/10.7554/eLife.17047.011 Image collected and cropped by CiteAb from the following publication (https://elifesciences.org/articles/17047), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human AKT by Simple Western
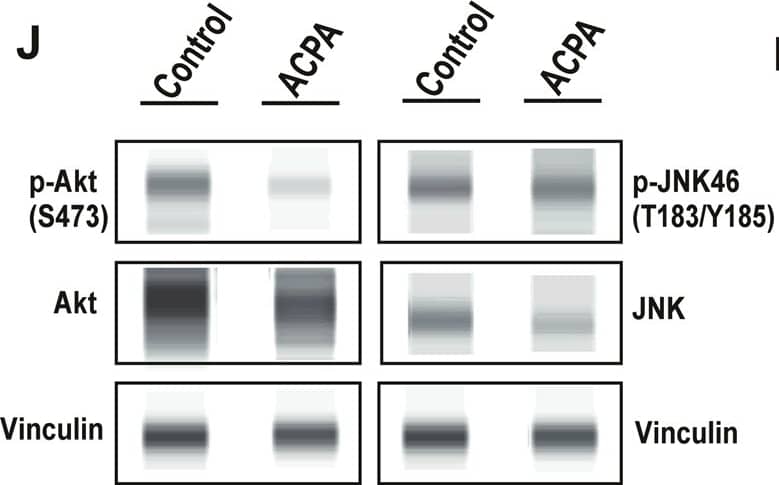
ACPA inhibits p-Akt, induces p-JNK and affects levels of specific metabolites in NSCLC lines.a Principal component analysis (PCA) score plot: Metabolomics profiling of control and ACPA-treated A549, H1299, H358, and H838 cells. b Changes in variable importance in projection (VIP) values for 19 metabolites in A549 cells. c, d, e Changes in VIP values for 20 metabolites in H1299, H358, and H838 cells. Significantly changed metabolites (*p < 0.05, indicated by arrows) were matched to apoptotic pathways. f, g, h, i Increase and decrease in several metabolites of ACPA-treated A549, H1299, H358, and H838 cells (*p < 0.05). j Simple Western showing total Akt, p-Akt (S473), total JNK46 and JNK54 and p-JNK46 and p-JNK54 (T183/Y185) in A549 cells at 24 hours after treatment with IC50 dose of ACPA. k Relative expression levels of Akt and p-Akt for control and ACPA-treated A549 cells after normalization by total vinculin protein. l Relative expression levels of JNK (46 and 54 kDa) and p-JNK for control and ACPA-treated A549 cells after normalization by total vinculin protein. *p < 0.05, Student’s t-test. All tests were done in quadruplicates. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/33431819), licensed under a CC-BY license. Not internally tested by R&D Systems.