SARS-CoV-2 Spike RBD LlaMABodyTM Bivalent VHH HuIgG2 Fusion Antibody
R&D Systems, part of Bio-Techne | Catalog # LMAB10731
Recombinant Monoclonal Antibody. Bivalent Llama VHH domain, Human IgG2 Fusion Antibody.

Key Product Details
Species Reactivity
SARS-CoV-2
Applications
Blockade of Receptor-ligand Interaction
Label
Unconjugated
Antibody Source
Recombinant Monoclonal Llama VHH domain Clone # L009.1.32H
Product Specifications
Immunogen
Human embryonic kidney cell HEK293-derived SARS-CoV-2 Spike RBD as immunogen for bivalent Llama VHH-Human IgG2 Fusion Antibody.
Arg319-Phe541
Accession # YP_009724390.1
Arg319-Phe541
Accession # YP_009724390.1
Specificity
Clone L009.1.32H is a bivalent Llama VHH-Human IgG2 Fusion Antibody that detects SARS-CoV-2 Spike RBD in direct ELISAs. Antibody construct is depicted below.SARS-CoV-2 Spike RBD SpecificLlama VHH domainLlama Hinge Human IgG2N-terminusC-terminus
Clonality
Monoclonal
Host
Llama
Isotype
VHH domain
Endotoxin Level
<0.10 EU per 1 μg of the antibody by the LAL method.
Scientific Data Images
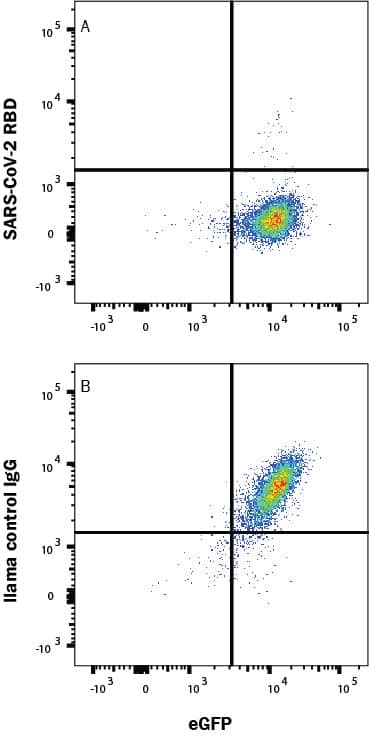
SARS-Cov-2 Spike 1 protein binding to ACE-2-transfected Human Cell Line is Blocked by SARS-Cov-2 Spike RBD Antibody.
In a functional flow cytometry test, Recombinant SARS-Cov-2 Spike 1 His-tagged protein (10522-CV) binds to HEK293 human embryonic kidney cell line transfected with recombinant human ACE-2 and eGFP. (A) Binding is completely blocked by 50 µg/mL of Llama Anti-SARS-Cov-2 Spike RBD Monoclonal Antibody (Catalog # LMAB10731) but not by (B) Llama IgG1 Control. Protein binding was detected with Mouse Anti-His APC-conjugated Monoclonal Antibody (IC050A). Staining was performed using our Staining Membrane-Associated Proteins protocol.Applications
Application
Recommended Usage
Blockade of Receptor-ligand Interaction
50 µg/mL
Sample: In a functional flow cytometry test, 50 μg/mL of Llama Anti-SARS-Cov-2 Spike RBD Antibody(Catalog # LMAB10731) will block the binding of Recombinant SARS-Cov-2 Spike 1 His-tagged protein (Catalog # 10522-CV) to HEK293 human embryonic kidney cell line transfected with recombinant human ACE-2.
Sample: In a functional flow cytometry test, 50 μg/mL of Llama Anti-SARS-Cov-2 Spike RBD Antibody(Catalog # LMAB10731) will block the binding of Recombinant SARS-Cov-2 Spike 1 His-tagged protein (Catalog # 10522-CV) to HEK293 human embryonic kidney cell line transfected with recombinant human ACE-2.
Formulation, Preparation, and Storage
Purification
Protein A or G purified from cell culture supernatant
Reconstitution
Reconstitute at 0.5 mg/mL in sterile PBS. For liquid material, refer to CoA for concentration.
Formulation
Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. *Small pack size (SP) is supplied either lyophilized or as a 0.2 µm filtered solution in PBS.
Shipping
Lyophilized product is shipped at ambient temperature. Liquid small pack size (-SP) is shipped with polar packs. Upon receipt, store immediately at the temperature recommended below.
Stability & Storage
Use a manual defrost freezer and avoid repeated freeze-thaw cycles.
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: Spike RBD
References
- Wu, F. et al. (2020) Nature 579:265.
- Tortorici, M.A. and D. Veesler (2019). Adv. Virus Res. 105:93.
- Bosch, B.J. et al. (2003). J. Virol. 77:8801.
- Belouzard, S. et al. (2009) Proc. Natl. Acad. Sci. 106:5871.
- Millet, J.K. and G. R. Whittaker (2015) Virus Res. 202:120.
- Yuan, Y. et al. (2017) Nat. Commun. 8:15092.
- Walls, A.C. et al. (2010) Cell 180:281.
- Jiang, S. et al. (2020) Trends. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Ortega, J.T. et al. (2020) EXCLI J. 19:410.
- Wrapp, D. et al. (2020) Science 367:1260.
- Tai, W. et al. (2020) Cell. Mol. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Okba, N. M. A. et al. (2020). Emerg. Infect. Dis. https://doi.org/10.3201/eid2607.200841.
- Wang, X. et al. (2020) https://doi.org/10.1038/s41423-020-0424-9.
- Wang, K. et al. (2020) bioRxiv https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1.
Long Name
Spike Receptor Binding Domain
Gene Symbol
S
UniProt
Additional Spike RBD Products
Product Specific Notices
For research use only
Loading...
Loading...
Loading...
Loading...