Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Protein
R&D Systems, part of Bio-Techne | Catalog # 10717-CV
N501Y Mutation found in UK, South African, and Brazilian Variants

Key Product Details
- R&D Systems HEK293-derived Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Protein (10717-CV)
- Quality control testing to verify active proteins with lot specific assays by in-house scientists
- All R&D Systems proteins are covered with a 100% guarantee
Source
HEK293
Accession #
Structure / Form
Disulfide-linked homodimer
Conjugate
Unconjugated
Applications
Bioactivity
Product Specifications
Source
Human embryonic kidney cell, HEK293-derived sars-cov-2 Spike RBD protein
| SARS-CoV-2 Spike RBD (Arg319-Phe541)(Asn501Tyr) Accession # YP_009724390.1 |
IEGRMD | Human IgG1 (Pro100-Lys330) |
| N-terminus | C-terminus |
Purity
>95%, by SDS-PAGE visualized with Silver Staining and quantitative densitometry by Coomassie® Blue Staining.
Endotoxin Level
<0.10 EU per 1 μg of the protein by the LAL method.
N-terminal Sequence Analysis
Arg319
Predicted Molecular Mass
52 kDa
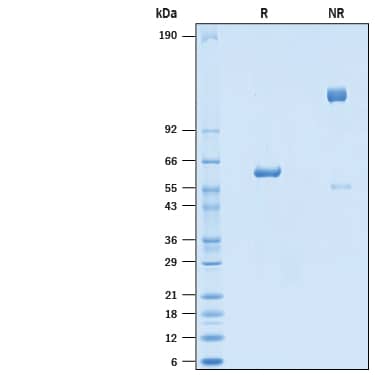
SDS-PAGE
55-65 kDa, under reducing conditions
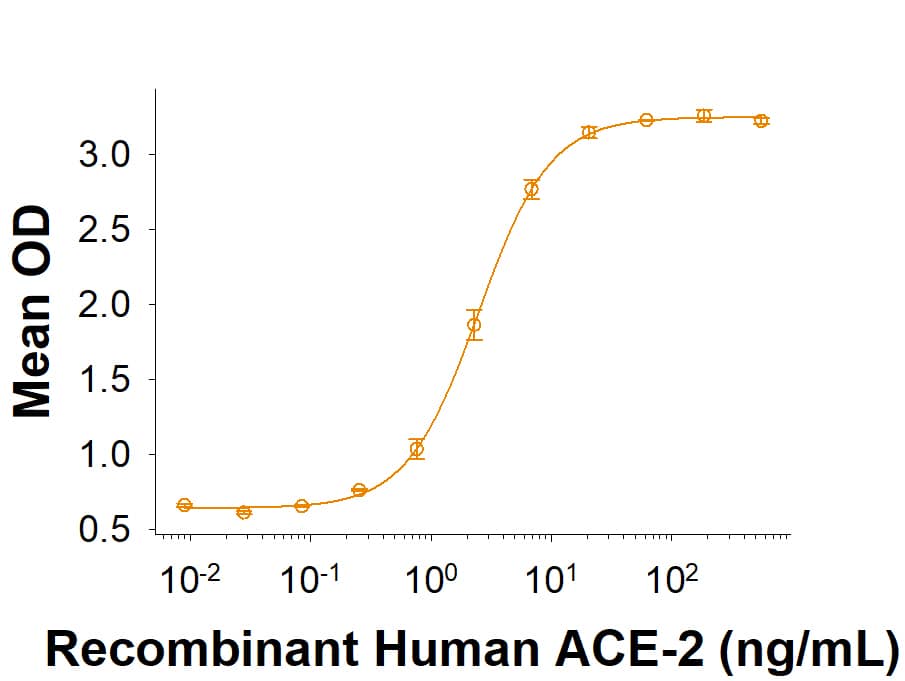
Activity
Measured by its binding ability in a functional ELISA with Recombinant
Human ACE-2 His-tag
(Catalog #
933-ZN).
Scientific Data Images for Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Protein
Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Chimera Protein Bioactivity
Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Chimera (Catalog # 10717-CV) binds Recombinant Human ACE-2 His-tag (933-ZN) in a functional ELISA.Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Chimera Protein SDS PAGE.
2 μg/lane of Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Chimera (Catalog # 10717-CV) was resolved with SDS-PAGE under reducing (R) and non-reducing (NR) conditions and visualized by Coomassie® Blue staining, showing bands at 55-65 kDa and 100-120 kDa, respectively.Binding of ACE-2 to SARS-CoV-2 Spike RBD N501Y Fc by surface plasmon resonance (SPR).
Recombinant SARS-CoV-2 Spike RBD N501Y Fc was immobilized on a Biacore Sensor Chip CM5, and binding to recombinant human ACE-2 (933-ZN) was measured at a concentration range between 0.18 nM and 47.2 nM. The double-referenced sensorgram was fit to a 1:1 binding model to determine the binding kinetics and affinity, with an affinity constant of KD=0.9196 nM.Formulation, Preparation and Storage
10717-CV
| Formulation | Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. |
| Reconstitution | Reconstitute at 500 μg/mL in PBS. |
| Shipping | The product is shipped at ambient temperature. Upon receipt, store it immediately at the temperature recommended below. |
| Stability & Storage | Use a manual defrost freezer and avoid repeated freeze-thaw cycles.
|
Background: Spike RBD
References
- Wu, F. et al. (2020) Nature 579:265.
- Tortorici, M.A. and D. Veesler (2019) Adv. Virus Res. 105:93.
- Bosch, B.J. et al. (2003). J. Virol. 77:8801.
- Belouzard, S. et al. (2009) Proc. Natl. Acad. Sci. 106:5871.
- Millet, J.K. and G.R. Whittaker (2015) Virus Res. 202:120.
- Li, W. et al. (2003) Nature 426:450.
- Wong, S. K. et al. (2004) J. Biol. Chem. 279:3197.
- Kozlov, Max (2020) TheScientist https://www.the-scientist.com/news-opinion/new-sars-cov-2-variant-spreading-rapidly-in-uk-68292.
- Wise, J. (2020) B.M.J. 371:m4857.
- Tang, J.W. et al. (2020) J. Infect. doi: 10.1016/j.jinf.2020.12.024.
- Davies, N.G. (2020) medRxiv doi:10.1101/2020.12.24.20248822.
Long Name
Spike Receptor Binding Domain
Gene Symbol
S
UniProt
Additional Spike RBD Products
Product Documents for Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Protein
Product Specific Notices for Recombinant SARS-CoV-2 B.1.1.7 N501Y Spike RBD Fc Protein
For research use only
Loading...
Loading...
Loading...