Autophagy Signaling Pathway
Autophagy-related proteins (ATG) regulate this process in yeast and many ATG proteins are conserved in mammals. Individual ATG proteins and ATG-complexes support specific steps in the dynamic process of autophagy. The formation of a double membrane vesicle for the entrapment and delivery of cytosolic content to lysosomes is the hallmark of autophagy. Formation of the mature organelle or autolysosome involves a sequence of coordinated events. In each step, ATG proteins catalyze specific reactions critical for the maintenance of autophagic flux.
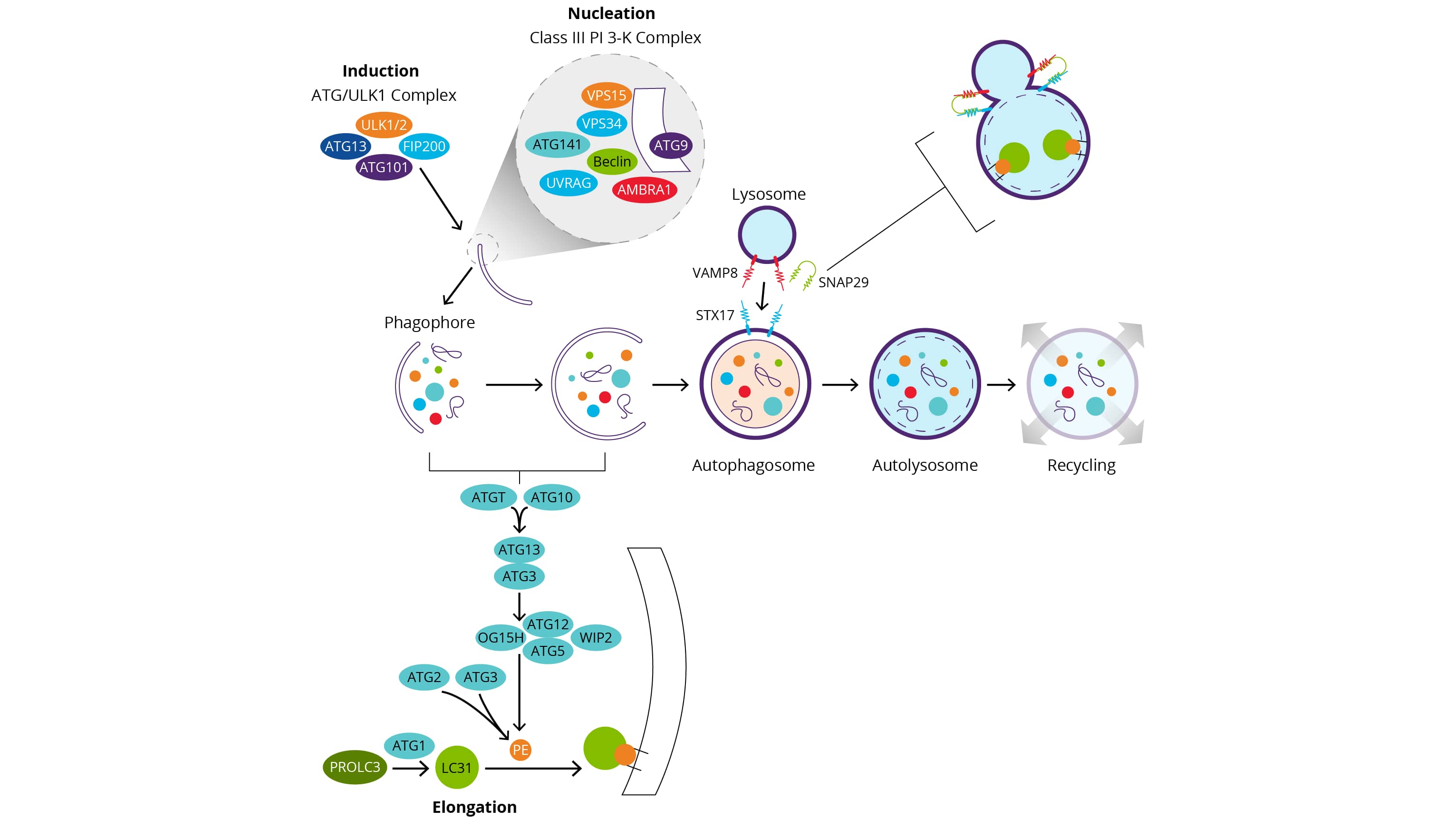
Induction
- May be triggered by starvation or reduced nutrient availability.
- Inhibition of mTOR kinase (mammalian target of rapamycin), a key regulator of nutrient signaling.
- Upon mTOR inhibition, decrease phosphorylation status of a protein complex including ULK1 (unc-51-like kinase 1), ATG13/ATG101 and FIP200/RB1CC1 (RB1-inducible coiled-coil 1), increases the activity of ULK1 and induces autophagy.
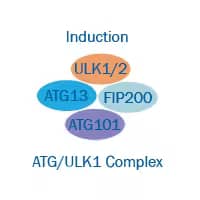
Nucleation, Phagophore Formation
- Release of Beclin1 and AMBRA1 from their inhibitor Bcl-2.
- Formation of the Ptdlns3K (Class lll phosphatidylinositol 3-kinase) complex including ATG14, VPS15 and VPS34 (vacuolar protein sorting).
- PtdIns3K complex phosphorylates lipids and gives rise to phosphatidylinositol-3-phosphate and recruits ATGs to the new membrane.
- The phagophore transmembrane protein ATG9 participates in the addition of membrane during elongation.
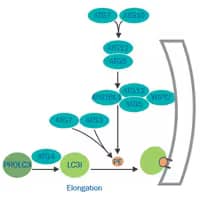
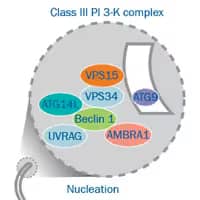
Elongation, Autophagosome Formation
- Mediated by two ubiquitin-like conjugation complexes: ATG7 and ATG10 act as E1 and E2 enzymes to conjugate ATG12 to ATG5.
- Atg12-Atgt5 conjugate with ATG16L1 (Atg16-like1) and promote the lipidation of LC3-II.
- ATG4, ATG7 (E1) and ATG3 (E2) process pro-LC3 to the final lipid conjugated form LC3-II.
- Phagophore’s membrane expands and closes onto itself sequestering the cytosolic content.
- All peripheral ATG proteins dissociate from the autophagosome and return to the cytosol.
- Lipidated LC3 remains associated with the autophagosome’s internal membrane.
- LC3-II interacts with p62/SQSTM1 inside the autophagosome.
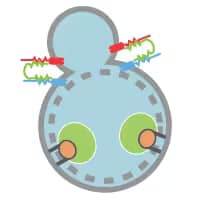
Fusion and Degradation
- SNARE protein STX17 (syntaxin 17 - blue) localizes to completed autophagosomes.
- STX17 interacts with SNAP29 - green and lysosomal VAMP8 - red and leads to autophagosome-lysosome fusion.
- Proton pumps and hydrolases are added to the autophagosome for formation of the autolysosome, cargo acidification and hydrolysis.
Selective Autophagy
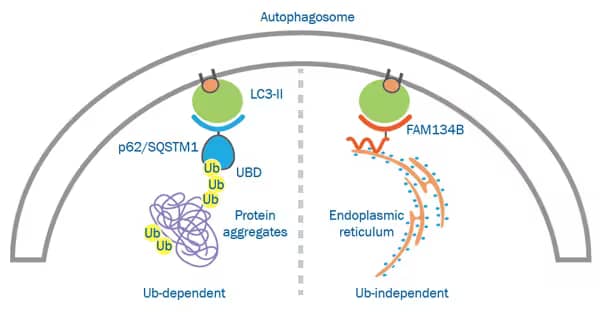
Cytosolic components are targeted for degradation in bulk or by selective autophagy. Selective autophagy pathways are broadly classified as ubiquitin-independent or -dependent and involve adaptor proteins and selective autophagy receptors that interact directly or via ubiquitin with cellular targets, respectively.
Autophagy Receptors
Ubiquitin-Dependent Autophagy: In ubiquitin-dependent autophagy, selective autophagy receptor proteins interact with their specific cargo via their ubiquitin binding domain (UBD). This selective autophagy pathway is believed to cooperate with the ubiquitin-proteasome system in the targeting and elimination of protein aggregates. Considerable overlap exists in the selectivity of the receptors.
Ubiquitin-Independent Autophagy: Receptors involved in this pathway recognize a variety of molecules as cargo including proteins, sugars and lipids. This type of selective autophagy was initially identified in yeast, but more recently specific pathways have been recognized in eukaryotes.
| Cytosolic Cargo | Ubiquitin-Dependent | Ubiquitin-Independent |
|---|---|---|
| Mitochondria | OPTN, NDP52, TAX1BP1, p62 | NIX/BNIP3, FUNDC1, ATG32 |
| Protein Aggregates | p62, NBR1, OPTN, TOLLIP | OPTN |
| Peroxisomes | NBR1, p62 | ATG30, ATG36 |
| Bacteria | p62, OPTN, NDP52, TAX1BP1 | Galectin-8/NDP52 |
| RNA granules | NDP52, p62 | |
| Proteasome | RPN10 | |
| Endoplasmic Reticulum | FAM134B | |
| Viruses | TRIM5α, SMURF1, p62 | |
| Nuclear Envelope | ATG39 |
LC3 Isoforms: LC3 and GABARAP Family Members
In mammals seven LC3 family members have been identified that belong to two main sub-families, LC3 and GABARAP. Different LC3 proteins may play specific roles in selective autophagy mechanisms.
| Sub-Family | Isoforms |
|---|---|
| LC3 | MAP1LC3A, MAP1LC3B, MAP1LC3B2, MAP1LC3C |
| GABARAP | GABARAP, GABARAPL1, GABARAPL2, GABARAPL3* |
Isoforms share 29-94% sequence identity. *Only subfamily member not involved in formation of autophagosomes.
Nuclear Regulation: Transcriptional Regulation
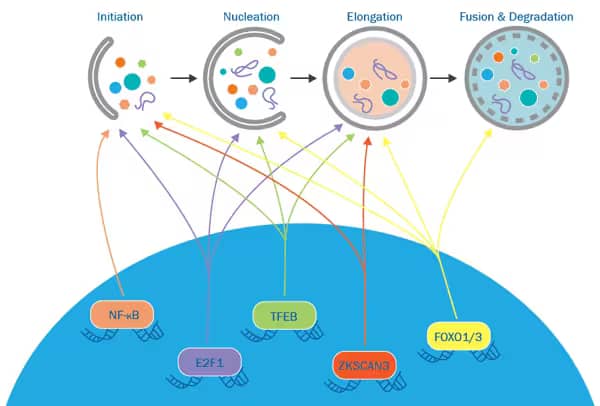
Various transcriptional regulatory axes have been identified to modulate autophagy. Translocation of transcription factors between the cytoplasm and the nuclei determines their function as modulators of autophagy. Each axis regulates autophagy via unique mechanisms.
- TFEB/ZKSCAN3 axis: Both transcription factors bind to overlapping promoter regions and regulate the expression of similar ATG genes.
- FOXO axis: FOXO1 is required for the transcriptional activation of autophagy genes by FOXO3.
- E2F1 and NF-kB axis: Inhibitory crosstalk between these two transcription factors is involved in the regulation of autophagy.
Epigenetic Control
Research in autophagy has predominantly centered on the cytoplasmic mechanisms that regulate this process. However, recent findings have identified intricate regulatory pathways involving DNA and histone-modifying enzymes that dynamically fine-tune the process of autophagy.
| Enzymatic Mechanism | Autophagy-related Genes Modulated | Associated Autophagic State |
|---|---|---|
| DNMT2 Hypermethylation | ATG5 and LC3 downregulated | Reduced autophagy |
| ESA1/RPD3 axis Acetylation/Deacetylation of H4 | Ribosomal protein upregulated; downregulated LC3 expression regulation | Reduced/Increased autophagy |
| G9A Methylation of H3K9 | LC3, WIPI1 and DOR downregulated | Reduced autophagy |
| hMOF/SIRT1 axis Acetylation/Deacetylation of H4K16 | Autophagy-related genes induced/ inhibited | Balance between cell death and survival |
| USP44 deubiquitination of H2B | Downregulated genes: regulation of NF-κB and biosynthetic process Upregulated genes: innate immunity and polyubiquitination | Increased autophagy |