Mitochondrial Markers
Common Mitochondrial Markers | Apoptosis | Oxidative Phosphorylation | Mitochondrial Dynamics | Additional Mitochondrial Markers

What are Mitochondria?
Mitochondria are compartmentalized, dual-membrane organelles that are responsible for most energy production in the cell. Mitochondria contain their own DNA and ribosomes. Their primary function is to convert energy from glucose into ATP via oxidative phosphorylation. Mitochondria also play an important role in the cellular stress response through processes such as autophagy, apoptosis, and hypoxia. Mitochondrial dysfunction is implicated in many human diseases including cancer, neurodegenerative disorders, and metabolic syndrome.
What are Mitochondrial Marker Antibodies Used For?
In response to stress or other biological cues, a protein’s subcellular distribution can change, indicative of the physiological state of the cell or tissue. Mitochondrial markers are used as well-established references when investigating the localization of a protein of interest and in microscopy furnishes additional details about mitochondrial structure and morphology. Mitochondrial markers also serve as quality controls. The purity of protein extracts derived from subcellular fractionation (Mitochondrial Isolation Kit [NBP2-29448]) can be examined in Western Blot to assess potential cross contamination of fractions (e.x. nucleus, cytosol, and mitochondria). Bio-Techne offers numerous highly published, well validated antibodies for mitochondrial markers along with bright, photostable, fluorescent conjugates such as Janelia Fluor® Dyes, for studying these complex, dynamic organelles. Janelia Fluor® Dyes are optimal for live cell imaging and super resolution microscopy, enabling superior detection and analysis of cellular targets.
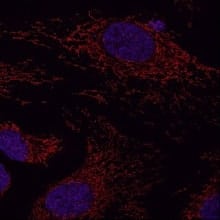
Image with super-resolution radial fluctuations microscopy analysis of outer mitochondrial membrane protein Tomm20 in HeLa cells. Tomm20 was detected using 1 µg/mL Rabbit anti-Tomm20 (NBP1-81556) and 1:500 JF646-conjugated anti-Rabbit secondary antibody (red) and counterstained with DAPI (blue). Scale bar: 10 μm. Experiment run by Jill Jenkins and images acquired at the Advanced Light Microscopy Core Facility, University of Colorado.
Apoptosis
Mitochondria play a crucial role in apoptosis, a form of programmed cell death, that is essential for establishing proper tissue homeostasis and is often dysregulated in cancer. The intrinsic/mitochondrial apoptosis pathway is initiated by factors including DNA damage and oxidative stress, leading to caspase activation and cell death. In mammals, apoptosis is a complex process largely regulated by the Bcl-2 family of proteins that are either pro-apoptotic (e.g. Bax, Bak) or anti-apoptotic (e.g. Bcl-2). In response to apoptotic stimuli, Bax/Bak become activated and Bax translocates to the mitochondria. The subsequent oligomerization of Bax/Bak promotes mitochondrial outer membrane permeabilization (MOMP), cytochrome c release and apoptosome formation. The resulting signaling cascade involving caspase-9 and caspase-3 activation ultimately leads to cell disassembly.
| Antibody Name | Location | Function |
| AIF (Apoptosis Inducing Factor) | Intermembrane space | Oxidoreductase that contributes to assembly of the respiratory chain and cell death. AIF deficiency leads to mitochondrial dysfunction. |
| APAF-1 | Cytoplasm (outside mitochondria) | Mediates formation of the apoptosme by binding cytochrome c following its release into the cytoplasm in the presence of ATP. |
| Cytochrome c | Intermembrane space | Role in electron transport and mediator of apoptosis. Released from mitochondria into cytoplasm and initiates apoptotic cascade and cell death. |
| PUMA | Cytoplasm; translocates to mitochondrial outer membrane | PUMA (p53 upregulated modulator of apoptosis) interacts with Bcl-2 family members after activation, promoting mitochondrial apoptosis and release of cytochrome c. |
| SMAC/Diablo | Intermembrane space | Released from mitochondria into the cytoplasm during apoptosis where it binds Inhibitor of apoptosis proteins (IAPs) and neutralizes their inhibitory action. |
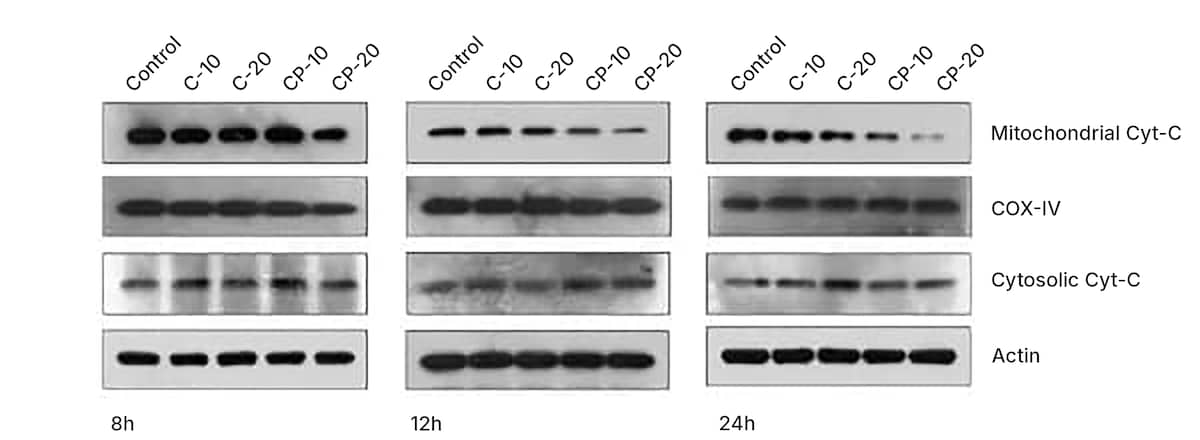
Western blots of apoptosis regulatory proteins, highlighting cytochrome c antibody (NB100-56503) (cytosolic and mitochondrial fractions). COX IV and β-actin were used as a loading control. HCT 116 cells were treated with 10 μM and 20 μM of Curcumin (C; NBP2-26243) and curcumin-PLGA (CP) conjugate for 6 h, 12 h and 24 h. Similarly, 0.8 μL DMSO (Catalog # 3176) was used as a vehicle control. Image collected and cropped from CiteAb from the following publication: (http://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC4334672&blobtype…) licensed under a CC-BY license.
Oxidative Phosphorylation
The most well-known function of mitochondria is energy production in the form of ATP. In the mitochondria, the citric acid cycle produces ATP and the reduction of coenzymes NAD+ and FAD+. During oxidation, the transfer of electrons from these reduced coenzymes involves a series of four respiratory chain (also known as the electron transport chain, ETC) protein complexes (Complex I, II, III, and IV), which creates a proton gradient across the inner mitochondrial membrane. These four complexes and the ATP synthase of the OXPHOS system are comprised of multiple subunits with over 90+ structural proteins and 30+ assembly factors
| Antibody Name | Location | Function |
|---|---|---|
| ATP5A | Inner membrane | Mitochondrial ATP synthase subunit that contributes to the ATP synthesis coupled proton transport. |
| ATP5F1 | Inner membrane | Mitochondrial ATP synthase subunit that catalyzes ATP from ADP by utilizing a proton gradient generated by the ETC. |
| BCS1L | Matrix | Chaperone protein involved in the assembly of Complex III of the ETC. |
| COX4 (Cytochrome c Oxidase Subunit IV isoform 1) | Inner membrane | Part of Complex IV in the ETC that transfers protons across the membrane and converts oxygen to water molecules. |
| COX5b | Matrix | Subunit of Cytochrome c oxidase complex that transfers electrons from cytochrome c to oxygen and helps generate the proton gradient across the inner membrane. |
| Glutamate Dehydrogenase | Matrix | Enzyme that catalyzes the reversible reaction between glutamate and alpha-ketoglutarate using NAD+ and NADP+. |
| NDUFC2 | Matrix | Subunit of NADH dehydrogenase (Complex I) which transfers electrons from NADH to the respiratory chain. |
| PDK4 | Matrix | Pyruvate dehydrogenase kinase isoform 4 (PDK4) inhibits the pyruvate dehydrogenase complex via phosphorylation, helping to regulate glucose metabolism. |
| Pyruvate Dehydrogenase | Matrix | Nuclear-encoded mitochondrial matrix multienzyme complex responsible for converting pyruvate to acetyl-CoA. |
| UQCRC1 | Matrix | Subunit of Complex III in the mitochondrial respiratory chain that catalyzes electron transfer from ubiquinol to cytochrome c. |
*Note: This table is not an exhaustive list of all mitochondrial proteins involved in oxidative phosphorylation.
Mitochondrial Dynamics
Mitochondrial dynamics refers to the ability of mitochondria to change shape, alter the inner and outer membrane structure, and exchange organelle components. The process of mitochondrial dynamics involves a coordination between fission and fusion events. Key components of the mitochondrial fission and fusion machinery include conserved GTPases in the dynamin family. Mitochondrial fission (mitochondrial fragmentation) is the process of generating multiple heterogeneous mitochondria from one mitochondrion. Conversely, mitochondrial fusion is the merging of two or more mitochondria to create a network.
Defective mitochondrial dynamics are a hallmark of many diseases, in particular for neurodegenerative disorders. A study by Wu et al. examined mitochondrial dysfunction in the context of Alzheimer’s disease (AD), finding altered mitochondrial morphology and significantly increased expression of key fusion proteins, including Mitofusin-1 (Mfn-1) (NBP1-71775) and Mitofusin-2 (Mfn-2). Wu et al. suggested excessive amyloid-beta (Aβ) accumulation leads to abnormal mitofusin expression and triggers mitochondrial dysfunction observed during AD pathogenesis.
| Antibody Name | Fission or Fusion | Location | Function |
|---|---|---|---|
| DRP1 (Dynamin-related protein 1) | Fission | Outer membrane | Functions in mitochondrial fission during apoptosis and required for proper cellular distribution of mitochondria. |
| Mitofusin 1 | Fusion | Outer membrane | Role in mitochondrial clustering, outer membrane fusion, and morphology. |
| Mitofusin 2 | Fusion | Outer membrane | Mitochondrial outer membrane fusion and a role in maintenance and operation of the mitochondrial network. |
| OPA1 (Optic Atrophy 1) | Fusion | Inner membrane | Similarity to dynamin-related GTPases with role in mitochondrial inner membrane fusion. |
Image with super-resolution radial fluctuations microscopy analysis of outer mitochondrial membrane protein Tomm20 in HeLa cells. Tomm20 was detected using 1 µg/mL Rabbit anti-Tomm20 (NBP1-81556) and 1:500 JF646-conjugated anti-Rabbit secondary antibody (red) and counterstained with DAPI (blue). Scale bar: 10 μm. Experiment run by Jill Jenkins and images acquired at the Advanced Light Microscopy Core Facility, University of Colorado.

Additional Mitochondrial Markers
Mitochondria have additional functions ranging from protein translation, preprotein and protein import and export, protein folding and assembly, and regulation of cell cycle and growth. HSP60, a well-known mitochondrial chaperonin, is responsible for regulating proper protein folding and assembly of imported proteins. Prohibitin, a ubiquitously expressed, pleiotropic protein, is involved in mitochondrial biogenesis, metabolism, and morphology, and, its dysregulation has been linked to aging, metabolic, proliferative, and neurodegenerative diseases.
| Antibody Name | Location | Function |
|---|---|---|
| Hexokinase 1 | Outer membrane | Phosphorylates glucose to produce glucose-6-phosphate and committing it to the glycolytic pathway. |
| HSP60 (Heat Shock Protein 60kDa) | Matrix | Protein folding and assembly of imported polypeptide. |
| HSP70 (Heat Shock Protein 70kDa) | Matrix | Role in protein folding, degradation, and association with presequence translocase-associated motor (PAM) to drive preprotein import. |
| Mitofilin | Inner membrane / Cristae | One of most abundant mitochondrial proteins. Role in maintenance of cristae morphology. |
| Prohibitin | Inner membrane | Show antiproliferative activity and has been proposed to play a role in normal cell cycle regulation, replicative senescence, cellular immortalization, and tumor suppression. |
| TIMM23 | Inner membrane | Subunit of the translocase of inner membrane 23 (TIM23) complex that functions to transport peptide-contain proteins across the inner membrane. |
| TOMM20 | Outer membrane | Together with TOM22 functions as the transit peptide receptor and facilitates the movement of preproteins into the TOM40 translocation pore. |
| VDAC1 | Outer membrane | Voltage dependent anion channel (VDAC) pore that allows entry and exit between the cytosol and mitochondria. Critical substrate for PINK/Parkin-mediated autophagy. |
*Note: This table is not an exhaustive list of all mitochondrial proteins involved in oxidative phosphorylation.
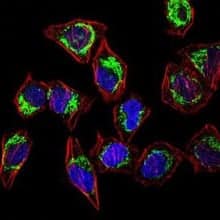
Confocal immunofluorescent analysis of HeLa cells staining with Rabbit Anti-Hsp60 Polyclonal antibody (NBP1-77397) at 1:100 followed by Alexa Fluor 488-conjugated Goat to rabbit IgG secondary antibody (green). Actin filaments were labeled with Alexa Fluor 568 phalloidin (red) and nuclei were stained with DAPI (blue).
Selected References
Bano, D., & Prehn, J. (2018). Apoptosis-Inducing Factor (AIF) in Physiology and Disease: The Tale of a Repented Natural Born Killer. EBioMedicine. https://doi.org/10.1016/j.ebiom.2018.03.016
Eisner, V., Picard, M & Hajnoczky, G. (2018) Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nature Cell Biology 20 755-765.
Javadov, S., Kozlov, A. V., & Camara, A. (2020). Mitochondria in Health and Diseases. Cells. https://doi.org/10.3390/cells9051177
Jeong, S. Y., & Seol, D. W. (2008). The role of mitochondria in apoptosis. BMB Reports. https://doi.org/10.5483/bmbrep.2008.41.1.011
Koopman, W. J., Distelmaier, F., Smeitink, J. A., & Willems, P. H. (2013). OXPHOS mutations and neurodegeneration. The EMBO Journal. https://doi.org/10.1038/emboj.2012.300
Lee, H., & Yoon, Y. (2016). Mitochondrial fission and fusion. Biochemical Society Transactions. https://doi.org/10.1042/BST20160129
Neupert W. (1997). Protein import into mitochondria. Annual Review of Biochemistry. https://doi.org/10.1146/annurev.biochem.66.1.863
Nunnari, J., & Suomalainen, A. (2012). Mitochondria: in sickness and in health. Cell. https://doi.org/10.1016/j.cell.2012.02.035
Pfanner, N., Warscheid, B., & Wiedemann, N. (2019). Mitochondrial proteins: from biogenesis to functional networks. Nature Reviews. Molecular Cell Biology. https://doi.org/10.1038/s41580-018-0092-0
Signorile, A., Sgaramella, G., Bellomo, F., & De Rasmo, D. (2019). Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells. https://doi.org/10.3390/cells8010071
Spinelli, J.B. & Haigis M.C. (2018) The multifaceted contributions of mitochondria to cellular metabolism. Nature Cell Biology 20 745-754.
Tilokani, L., Nagashima, S., Paupe, V., & Prudent, J. (2018). Mitochondrial dynamics: overview of molecular mechanisms. Essays in Biochemistry. https://doi.org/10.1042/EBC20170104
Wang, C., & Youle, R. J. (2009). The role of mitochondria in apoptosis*. Annual Review of Genetics. https://doi.org/10.1146/annurev-genet-102108-134850
Westermann B. (2010). Mitochondrial fusion and fission in cell life and death. Nature Reviews. Molecular cell biology. https://doi.org/10.1038/nrm3013
Wiedemann, N., & Pfanner, N. (2017). Mitochondrial Machineries for Protein Import and Assembly. Annual Review of Biochemistry, 86, 685–714. https://doi.org/10.1146/annurev-biochem-060815-014352
Wu, Z., Zhu, Y., Cao, X., Sun, S., & Zhao, B. (2014). Mitochondrial toxic effects of Aβ through mitofusins in the early pathogenesis of Alzheimer's disease. Molecular Neurobiology. https://doi.org/10.1007/s12035-014-8675-z
Yadav, K., Yadav, A., Vashistha, P., Pandey, V. P., & Dwivedi, U. N. (2019). Protein Misfolding Diseases and Therapeutic Approaches. Current Protein & Peptide Science. https://doi.org/10.2174/1389203720666190610092840
