Detection Methods for Epigenetics
Epigenetic modifications determine how the information encoded in DNA sequences is translated into specific phenotypes. A wide range of epigenetic marks and modifier enzymes shape the epigenetic code. Understanding the complex interactions between different epigenetic marks which often influence the activities of epigenetic writers, readers and erasers requires diverse technical approaches. A variety of methods may be applied which target the specific marks or modifiers to understand the underlying mechanisms.
Methylated DNA
Various methods may be employed for the detection of methylated CpG sequences. Selection of a specific method depends partly on the objectives of the study (e.g., quantifying the extent of methylation genome wide vs methylation of a known gene region).
| Uncovering new methylation sites | Extent of methylation of known gene targets |
|---|---|
| Bisulfite conversion- followed by sequencing or Microarray analysis | Bisulfite conversion-followed by qPCR or PCR and sequencing |
| Genome wide methylation quantification- HPLC-UV, LC-MS/MS and ELISA | DNA enzyme digest based on target sequence- followed by qPCR or PCR and sequencing |
- Bisulfite conversion: In this method, a DNA sample is treated with sodium bisulfite resulting in the deamination of unmethylated cytosine to uracil and allowing the distinction between cytosine and methylated cytosine via sequencing and Next Generation Sequencing approaches.
- DNA enzyme digest: This method is based on the use of DNA endonucleases which do not cut methylated DNA. Digestion of specific DNA target sequences by these enzymes generates DNA fragments of different lengths which may be sequenced to determine the extent of methylation.
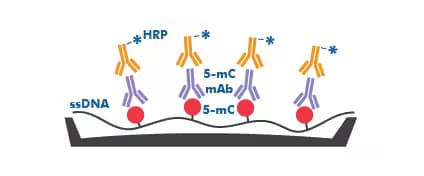
ELISA for detection of 5-methylated cytosine: Non-species specific 5-MethylCytosine ELISA Kit (Colorimetric) [NBP2-62131]: The workflow for the 5-Methylcytosine DNA ELISA kit utilizes the indirect ELISA methodology where denatured, single-stranded DNA (ssDNA) samples are coated on the plate well surfaces and a 5-mC mAb and conjugate HRP-Ab are added to the wells.
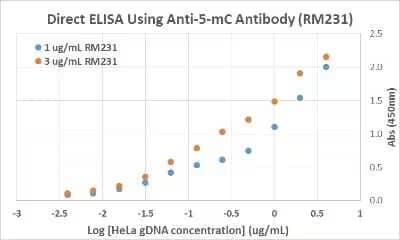
5-MethylCytosine Antibody (231) [NBP2-61470]: ELISA analysis of HeLa cell genomic DNA. The plate was directly coated with different concentrations of genomic DNA isolated from HeLa cells. 1 μg/mL or 3 μg/mL was used for the primary antibody, and an HRP conjugated anti-rabbit IgG as the secondary antibody.
Histone Modification
Chromatin post-translational modifications have been predominantly identified within the amino-terminal “tail” domain of histones. Modified histones influence chromatin’s structure and its interaction with readers, which in turn introduce additional modifications. The specific array of posttranslational modifications executed by the activities of writers, erasers and readers directly shape the expression of genes via different mechanisms.
To understand the role of histone modifications in regulating gene expression several methods are necessary to:
- Detect histone modifications (type and abundance): Immunoblot analysis including dot blots and WB are used to validate antibody specificity or to asses histone mark abundance. For most applications, WB analysis relies on the use of whole cell lysates, prepared by standard procedures with SDS Laemmli sample buffer. Some applications require the use of samples enriched for histone proteins which depend on nuclei or chromatin isolation
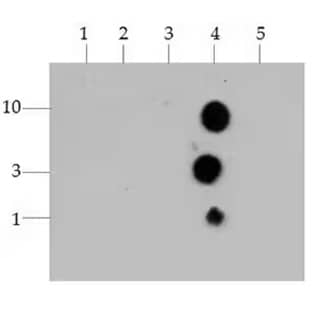
Histone H3 [Trimethyl Lys4] Antibody [NB21-1023]: Dot Blot analysis of Histone H3 K4me3 using the peptides listed at 10, 3, and 1 picomoles(pM)of peptide.
- K4
- K4-KMe1
- K4-KMe2
- K4-KMe3
- K23-KMe3
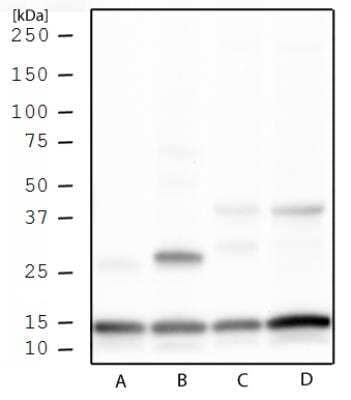
Histone H3 [Trimethyl Lys4] Antibody [NB21-1023]: Western blot analysis of Histone H3 K4me3 in HeLa histone prep (A), NIH 3T3 histone prep (B), HeLa whole cell extract (C), and NIH 3T3 whole cell extract (D) using Histone H3 [Trimethyl Lys4] antibody (NB21-1023) at 1 μg/mL.
- Identify modified-histone interacting proteins: Immunoprecipitation and WB analyses are used in combination for the identification of histone interacting proteins. Co-immunoprecipitation of histone-binding protein complexes requires the enzymatic processing of chromatin to generate single nucleosome units. Antibodies specific to the chromatin binding protein of interest are then used to pull down the complex. Subsequent WB analysis with antibodies to unmodified and modified histones allows the identification of the exact molecular nature of the complex.

Explore our range of multi-assay validated Epi-Plus antibodies for modified histones.
- Determine the genomic location of histone marks: Chromatin immunoprecipitation (ChIP) is used in combination with PCR, next generation sequencing (NGS) or microarray strategies to map the location and determine the abundance of modified histones or variants in the genome.
PCR: abundance of modified histones in targeted sequences
NGS (ChIP-seq): abundance of modified histones across the genome
Microarray (ChIP—chip): abundance of modified histones across the genome