by Victoria Osinski
What are PAMPs and DAMPs?
Inflammation results from tissue damage or infection, and the response can be beneficial or harmful depending on the type and duration of stimulation. The sources, structures, and abundance of these stimuli are highly variable and stimulate immune responses in context-specific ways. One major category of inflammatory stimuli includes the families of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs).1,2 These patterns are found in molecular components of bacterial cell walls, DNA, lipoproteins, carbohydrates, and other structures. While many DAMPs and PAMPs have been identified, there is considerable room for a deeper understanding of how they trigger inflammation.
PAMPs vs DAMPs: What’s the Difference?
PAMPs are derived from microorganisms and thus drive inflammation in response to infections.2 One well-known PAMP is lipopolysaccharide (LPS), which is found on the outer cell wall of gram-negative bacteria.3 DAMPs are derived from host cells including tumor cells, dead or dying cells, or products released from cells in response to signals such as hypoxia. Because they are derived from host materials, DAMPs induce what’s known as sterile inflammatory responses. DAMPs are often created or exposed in environments of trauma, ischemia, or tissue damage and do not require pathogenic infection.2,4 These environments develop in settings such as myocardial infarction, cancer, autoimmune disease, and atherosclerosis.5
Pattern Recognition Receptors: Signaling Downstream of PAMPs and DAMPs
PAMPs and DAMPs bind to pattern recognition receptors, which include Toll-like receptors (TLRs), cytoplasmic NOD-like receptors (NLRs), intracellular retinoic acid-inducible gene-I acid-inducible-like receptors (RLRs), transmembrane C-type lectin receptors, and Absent in Melanoma 2-like receptors (AIM2).3,5 Cell types expressing pattern recognition receptors include innate immune cells (e.g. macrophages, monocytes, dendritic cells, and mast cells) but also non-immune cells such as epithelial cells and fibroblasts.1,2 Pattern recognition receptor-ligand binding and their concomitant conformational changes trigger a cascade of downstream signaling that result in transcriptional changes as well as post-translational modifications.3 Broadly, pattern recognition receptor engagement results in signals that prompt leukocyte recruitment.3
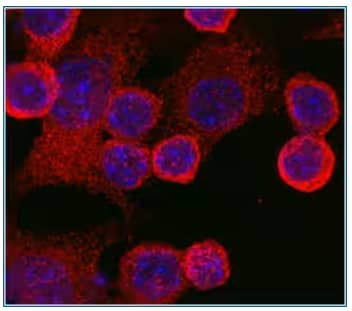
TLR4 was detected in immersion fixed RAW 264.7 mouse monocyte/macrophage cells using TLR4 Antibody (Catalog # MAB2759) at 10 µg/mL for 3 hours at room temperature. Cells were stained using the NorthernLights™ 557-conjugated Secondary Antibody (red; Catalog # NL013) and counterstained with DAPI (blue; Catalog # 5748). Specific TLR4 staining was localized to the cell surface.
Pattern Recognition Receptor Responses are Context-Dependent
Pattern recognition receptors are capable of recognizing a variety of molecular patterns, which in turn induce a receptor-dependent response. A single pattern recognition receptor can recognize multiple PAMPs and DAMPs, and the structural and molecular mechanisms mediating how this happens are still being studied.5 Furthermore, simultaneous signaling within the same cell can modulate downstream responses to pattern recognition receptor engagement. For example, cytokines can stimulate downstream signaling that may be complementary, amplifying, or inhibitory to pattern recognition receptor signaling pathways.1 Such complexities make the study of PAMP- and DAMP-induced inflammatory responses complicated but quite fascinating.

Victoria (Tori) Osinski, Doctoral Candidate
University of Virginia
Victoria studies cellular mechanisms regulating vascular growth during peripheral artery disease and obesity.
Additional Resources for PAMPs and DAMPs
Pattern Recognition Receptors on Endothelial Cells
Pattern Recognition Receptors at R&D Systems
Pattern Recognition Receptors Brochure at R&D Systems
Toll-like Receptors (TLRs) Ligands
Innate Immunity Cell Identity Markers
Signaling in Innate Immunity Poster
C-type Lectin Receptors at R&D Systems
A Guide to Human and Mouse Immune Cell Characterization
The Complex Biology of Macrophages: Origins, Functions, & Activation States Poster at R&D Systems
-
Newton, K. and Dixit, V.M. (2012) Signaling in Innate Immunity and Inflammation. Cold Spring Harb. Perspect. Biol.
-
Tang, D. et al. (2012) PAMPs and DAMPs: Signal 0s that Spur Autophagy and Immunity. Immunol. Rev. 249:158-175.
-
Mogensen, T.H. (2009) Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 22:240-273.
-
Bianchi, M.E. (2007) DAMPs, PAMPs and Alarmins: All We Need to Know About Danger. J. Leukoc. Biol. 81:1-5.
-
Schaefer, L. (2014) Complexity of Danger: The Diverse Nature of Damage-associated Molecular Patterns. J. Biol. Chem. 289:35237-35245.