Toll-like Receptors (TLRs)
The Toll-like receptors (TLRs) belong to a family of innate immune receptors known as pattern recognition receptors (PRRs), which includes Nod-like receptors (NLRs) and RIG-I like receptors (RLRs). Ten TLRs (TLR1-10) have been identified in humans and 13 have been identified in mice (TLR1-13). The TLRs recognize pathogen-associated molecular patterns (PAMPs) to activate innate immunity and inflammatory cascades, functioning as the first line of defense against foreign pathogens. These receptors can also bind damage-associated molecular patterns (DAMPs) to activate innate immune responses to “sterile” stimuli such as apoptotic cells and extracellular DNA. DAMPs are released following environmental stimuli such as hypoxia, in which cell death and cell stress signals are prevalent. Activation of inflammatory responses through TLR signaling allows tissue repair processes. However, abnormal TLR signaling has been shown to play roles in many diseases including cancer, sepsis, allergy, rheumatoid arthritis, and ischemia-reperfusion injury.
What is the Difference Between PAMPS and DAMPS?
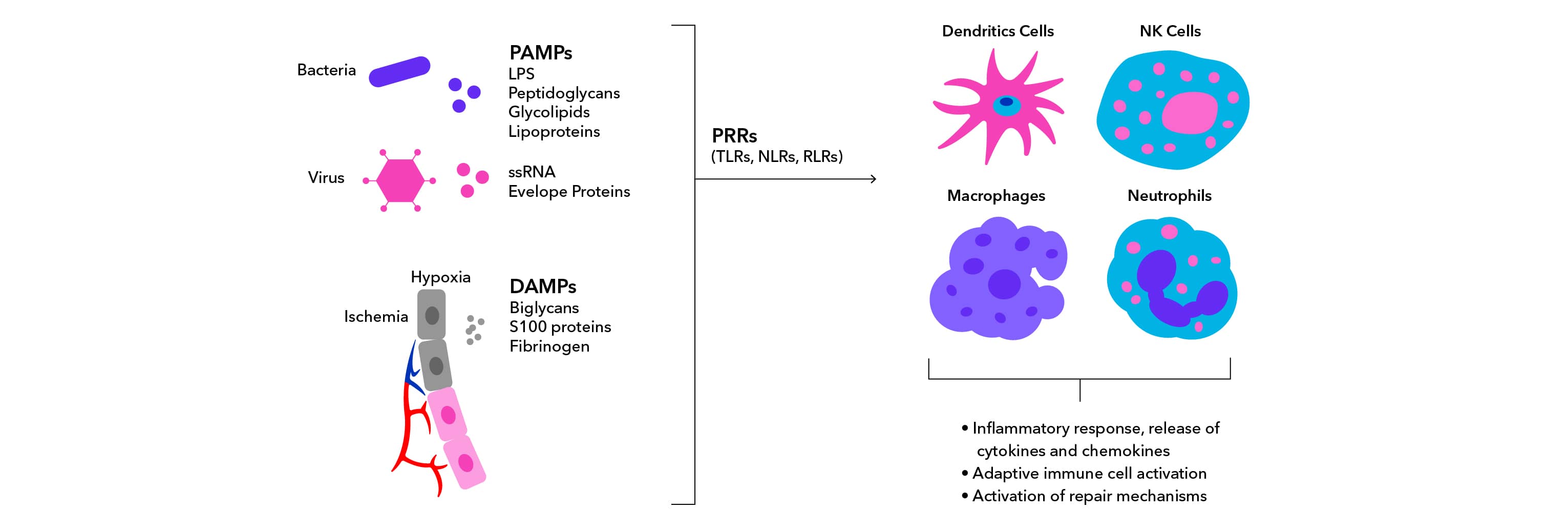
PAMPs are derived from micro-organisms and drive inflammation in response to infections. One well-known PAMP is lipopolysaccharide (LPS), which is found on the outer cell wall of gram-negative bacteria. DAMPs are derived from host cells including tumor cells, dead or dying cells, or products released from cells in response to stress such as hypoxia. Because they are derived from host materials and do not require pathogenic infection, DAMPs induce sterile inflammatory responses. DAMPs are often produced or exposed in environments of trauma, ischemia, or tissue damage in association with myocardial infarction, cancer, autoimmune disease, and atherosclerosis. Interaction of PAMPs and DAMPs with PRRs leads to activation of innate immunity and inflammatory responses.
TLR Structure
TLRs are type I transmembrane proteins consisting of several structural domains:
1) a leucine-rich repeats (LRRs) motif
2) a transmembrane domain
3) a cytoplasmic Toll/IL-1 receptor (TIR) domain
These receptors are found either on the plasma membrane or inside the cell associated with endosomal membranes. The LRR motif is located across the membrane from the TIR domain and is responsible for recognizing PAMPs and DAMPs. The TIR motif initiates signaling cascades through interactions with other signal transduction adaptors such as Myeloid differentiation factor 88 (MyD88) and TIR domain-containing adaptor inducing IFN-beta (TRIF).
TLR Signaling
TLR signaling activation can be initiated in multiple ways depending on the receptor, stimuli, and additional external and internal signals. One such activation is a MyD88 dependent mechanism, by which the TIR domain associates with the adaptor molecule MyD88 and MAL/TIRAP (TIR domain-containing adaptor protein). IL-1R-associated kinases IRAK1 and IRAK4 bind MyD88, activating downstream MAPKK and IKK signaling to induce nuclear translocation of transcription factors AP-1 and NFkB, respectively. TLR signaling can also be activated through a MyD88 independent mechanism, via the activation of TRIF and TNF receptor-associated factor 3 (TRAF-3). An additional molecular adaptor known as Sterile alpha and armadillo-motif-containing protein (SARM), can inhibit TLR signaling through interactions with both TRIF and MyD88.
Toll-like Receptors Function
TLR signaling can induce a variety of functions in cells, which are dependent on the specific receptor, cell type, and other co-stimulatory signals involved. TLRs can stimulate expression of pro-inflammatory cytokines, proliferation and survival mechanisms, and facilitate communication between innate and adaptive immunity. Broadly, TLR induced signaling promotes gene expression via master transcriptional regulators AP-1, NFkB, CREB, and IRFs. One exception to this pattern of signaling is observed with TLR10, which is anti-inflammatory. TLR10 inhibits TLR2 signaling and promotes expression of IL-1ra .
Explore TLR function with TLR Ligands and TLR Inhibitors.
| Ligands | Adaptor protein(s) | Cellular localization | Cell type expression | |
|---|---|---|---|---|
| TLR1 |
PAMPs Bacterial lipoproteins |
MyD88/MAL | Cell surface |
B Cell T cell Dendritic Cell Macrophage Granulocyte |
| TLR2 |
PAMPs Lipoteichoic acid (LTA) Peptidoglycan (PGN) Bacterial lipoproteins DAMPs Heat shock proteins (HSPs) |
MyD88/MAL | Cell surface |
Dendritic Cell Macrophage Granulocyte Endothelial cells Cardiomyocytes |
| TLR3 |
PAMPs Double-stranded RNA DAMPs mRNA |
TRIF | Endosome |
T Cell Dendritic Cell Macrophage |
| TLR4 |
PAMPs DAMPs |
Cell surface |
B Cell T cell Dendritic Cell Macrophage Granulocyte |
|
| TLR5 |
PAMPs |
MyD88/TRIF | Cell surface |
Dendritic Cell Macrophage Granulocyte |
| TLR6 |
PAMPs Lipoteichoic acid (LTA) Diacyl lipoproteins DAMPs |
MyD88/MAL | Cell surface |
B Cell Dendritic Cell Macrophage |
| TLR7 |
PAMPs Imidazoquinolines (Synthetic) ssRNA DAMPs |
MyD88 | Endosome |
B Cell Dendritic Cell Macrophage |
| TLR8 |
PAMPs Imidazoquinolines (Synthetic) ssRNA DAMPs |
MyD88 | Endosome |
Dendritic Cell Macrophage |
| TLR9 |
PAMPs Bacterial DNA with Unmethylated CpG DAMPs Nuclear DNA Mitochondrial DNA |
MyD88 | Endosome |
B Cell T cell Dendritic Cell Macrophage |
| TLR10 (human) |
PAMPs PAM3CSK4 (Synthetic) |
Unknown | Unknown |
B Cell T cell Dendritic Cell Macrophage |
| TLR11 (mouse) |
PAMPs Profilin
|
MyD88 | Endosome | Macrophages |
| TLR12 (mouse) |
PAMPs Profilin |
MyD88 | Endosome |
Dendritic Cells Macrophages |
| TLR13 (mouse) |
PAMPs 23S Ribosomal RNA |
MyD88 | Endosome |
Macrophages Dendritic Cells |
Content developed by Victoria Osinski.
Carlsson, E., Ding, J. L., & Byrne, B. (2016). SARM modulates MyD88-mediated TLR activation through BB-loop dependent TIR-TIR interactions. Biochimica et Biophysica Acta - Molecular Cell Research. https://doi.org/10.1016/j.bbamcr.2015.11.021
Goulopoulou, S., McCarthy, C. G., & Clinton Webb, R. (2016). Toll-like receptors in the vascular system: Sensing the dangers within. Pharmacological Reviews. https://doi.org/10.1124/pr.114.010090
Hatai, H., Lepelley, A., Zeng, W., Hayden, M. S., & Ghosh, S. (2016). Toll-like receptor 11 (TLR11) Interacts with flagellin and profilin through disparate mechanisms. PLoS ONE. https://doi.org/10.1371/journal.pone.0148987
Jiang, S., Li, X., Hess, N. J., Guan, Y., & Tapping, R. I. (2016). TLR10 Is a Negative Regulator of Both MyD88-Dependent and -Independent TLR Signaling. The Journal of Immunology. https://doi.org/10.4049/jimmunol.1502599
Li, X. D., & Chen, Z. J. (2012). Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. ELife. https://doi.org/10.7554/eLife.00102
Nie, L., Cai, S.-Y., Shao, J.-Z., & Chen, J. (2018). Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals. Frontiers in Immunology. https://doi.org/10.3389/fimmu.2018.01523
O’Neill, L. A. J., Bryant, C. E., & Doyle, S. L. (2009). Therapeutic targeting of toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacological Reviews. https://doi.org/10.1124/pr.109.001073
Oosting, M., Cheng, S. C., Bolscher, J. M., Vestering-Stenger, R., Plantinga, T. S., Verschueren, I. C., … Joosten, L. A. B. (2014). Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.1410293111
Midwood, K. S., & Piccinini, A. M. (2010). DAMPening inflammation by modulating TLR signalling. Mediators of Inflammation. https://doi.org/10.1155/2010/672395
Roh, J. S., & Sohn, D. H. (2018). Damage-associated molecular patterns in inflammatory diseases. Immune Network. https://doi.org/10.4110/in.2018.18.e27
Yang, D., Han, Z., & Oppenheim, J. J. (2017). Alarmins and immunity. Immunological Reviews. https://doi.org/10.1111/imr.12577
