By Christina Towers, PhD
Autophagy is a stress-induced cellular recycling process that plays an important physiological role in many diseases. It is induced by a variety of stimuli, both intracellular and extracellular, and is a tightly regulated process with over 30 core autophagy proteins necessary for efficient autophagosome formation and fusion with lysosomes to facilitate degradation1. One of the most common ways to monitor autophagy is by measuring the protein levels of LC3-II, which is incorporated into autophagosomes and then degraded in the lysosome. There are many different ways to measure LC3 protein levels, including Western blot and immunofluorescent microscopy2. These methods, however, are complicated by the fact that LC3 levels increase with autophagy induction due to increased autophagosomal formation but also decrease as these forming autophagosomes are turned over, and the only way to accurately monitor LC3 levels is with a flux assay using an autophagy inhibitor like Bafilomycin-A1 or Chloroquine.
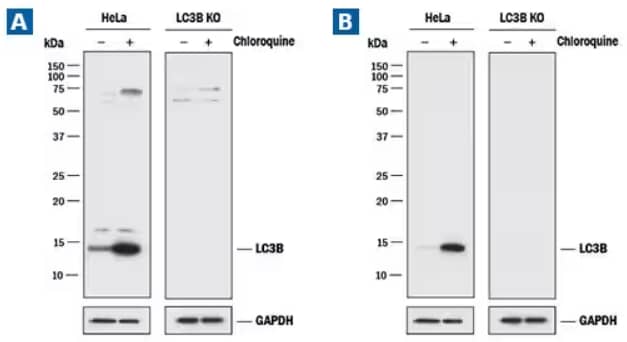
LC3B Knockout Validation. Lysates of HeLa cell line and LC3B knockout HeLa cell line (KO) untreated (-) or treated (+) with 50 μM Chloroquine (Catalog # 4109) for 18 hours. Polyvinylidene difluoride (PVDF) membrane was probed with (A) 0.5 μg/mL of Rabbit LC3B Polyclonal Antibody (Catalog # NB100-2220) followed by HRP-conjugated Anti-Rabbit IgG Secondary Antibody (Catalog #HAF008)or (B) 2.0 μg/mL of Rabbit Anti-Human/Mouse/Rat LC3B Monoclonal Antibody (1251A) (Catalog # NBP2-46892) followed by HRP-conjugated Anti-Rabbit IgG Secondary Antibody (Catalog #HAF008). A specific band was detected for LC3B at approximately 15 kDa in the parental HeLa cell line, but is not detectable in the knockout HeLa cell line. GAPDH is shown as a loading control. This experiment was conducted under reducing conditions.
Tandem constructs have been designed that couple pH-stable fluorophores like RFP or mCherry with the pH sensitive fluorophore, GFP, to monitor LC3 turn over in the lysosome. Attempts to quantify LC3 foci with microscopy can introduce experimenter bias, so these flux assays are qualitative at best.
Tandem mCherry-GFP-LC3 and Ratiometric Flow Cytometry
A more quantitative assay to measure autophagy couples the tandem mCherry-GFP-LC3 construct with ratiometric flow cytometry3. Images from a recent publication4 (Panels a and b) exemplify how this assay can be used to detect small changes in autophagic flux in cultured cells at both baseline and after treatment.
(a) To perform this assay, cultured cells with stable expression of mCherry-GFP-LC3 were treated with an experimental stimulus and then harvested for flow cytometry. The necessary controls include cells treated with Bafilomycin-A1 for 24 hours to completely inhibit autophagic flux, which will be used to set the flow cytometry gates. (b) Cells starved of amino acids and glucose for 4-24 hours serve as a positive control for induced autophagy, and unstained cells are used to eliminate low expressing cells from analysis.
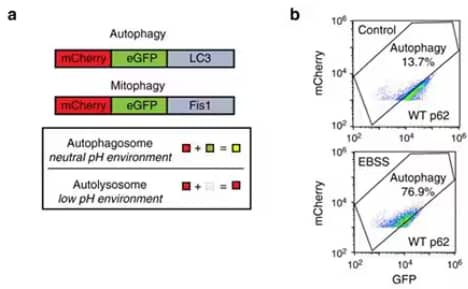
Section from figure 4 in recent publication by Zhang et al. (2018) (www.nature.com/articles/s41467-018-06878-8). Panels (c) to (g) are not shown for simplicity. For license, refer to creativecommons.org/licenses/by/4.0/.
Forward scatter and side scatter are used to gate live and single cells. An angled rectangular "autophagy" gate can then be set based on the Bafilomycin-A1-treated cells, such that 5-10% of cells score positive in this condition, and 50-80% of the cells shift left into this gate after starvation. Flow cytometry software like Flow Jo can run a ratiometric calculation as a derived parameter, where autophagic flux is the mCherry signal divided by the GFP signal. This assay is simple and quick with a 1-2 hr process time after treatment and is the most quantitative way to measure autophagic flux.

Christina Towers, PhD
University of Colorado (AMC)
Dr Towers studies the roles of autophagy, apoptosis and cell death in cancer.
-
Towers, C. G. & Thorburn, A. (2016) Therapeutic targeting of autophagy. EBioMedicine 14:15-23. PMID: 28029600.
-
Klionsky, D. J. et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1-222. PMID: 26799652.
-
Gump, J. M. & Thorburn, A. (2014) Sorting cells for basal and induced autophagic flux by quantitative ratiometric flow cytometry. Autophagy 10:1327-1334. PMID: 24915460.
-
Zhang, Y. et al. (2018) ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat Commun 9:4373. PMID: 30349045.