Hypoxia and HIFs - Frequently Asked Questions
|
Find scientific technical answers on frequently asked questions (FAQs) relating to:
|
||
|
Frequently asked questions (FAQs) on: hypoxia signaling experiments; hypoxia markers; HIF-1 alpha, HIF-2 alpha, HIF-1 beta, and HIF-3 alpha; hypoxia target genes; IHC, ICC, Flow, and WB troubleshooting tips for HIF-1/HIF-2 alpha analysis; positive controls, inhibitors and inducers for HIF-1/-2 alpha and more. What is Hypoxia? How is it relevant to life sciences? What are some key players in hypoxia signaling? |
Some of the major target genes of HIFs are listed below:
| Angiogenesis | EMT & Stemness | Invasion/Metastasis | Metabolism | Proliferation & Survival | pH Control & Misc |
|---|---|---|---|---|---|
| VEGFA VEGFR1 VEGFR2 PAI1 ANG1 ANG2 PDGFB TIE2 NOS2 |
ID2 SNAI1 SNAI2 TGF alpha ZEB1 OCT4 JARID1B Vimentin Twist-1 TERT NOTCH |
c-Met SDF1 UPAR Vimentin E-Cadherin KRT18 MMP2 MMP9 FN1 COL5A1 |
GLUT1 GLUT3 HK1 HK2 GAPDH LDHA PDK1 PKM2 ENO1 PGK1 |
Survivin Cyclin G2 IGF2 IGFBP2 CDKN1A CCND1 EPO TGFB3 Transferrin TfR BNIP3 |
CA9 CA12 DDIT4 AK1 HO1 PAI1 CITED2 Nur77 DEC1 DEC2 CD73 |
What is the significance of HIF-1 alpha in hypoxia?
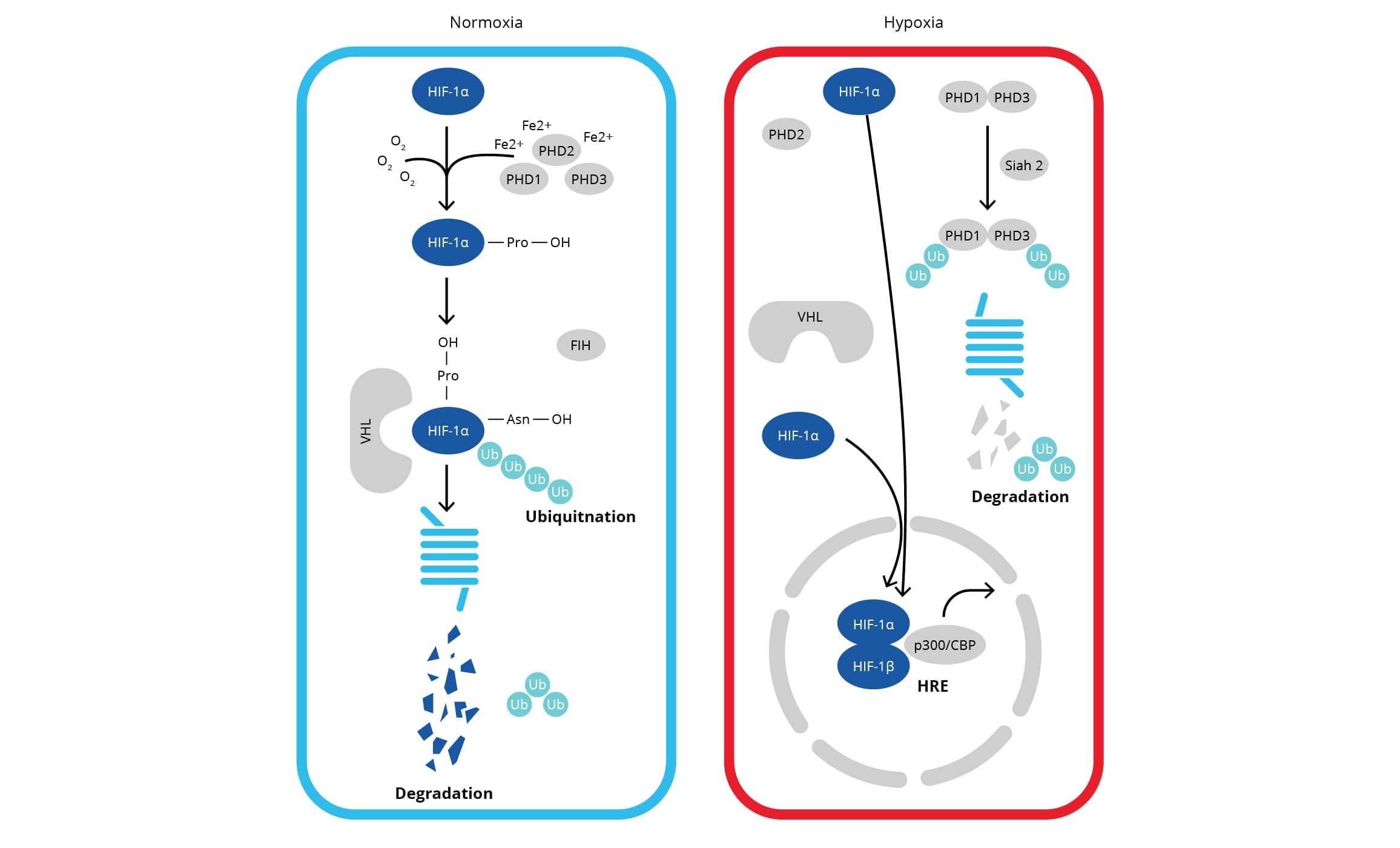
HIF1 (hypoxia-inducible factor 1) is a heterodimeric transcription factor complex which is central to the cellular response to hypoxia. HIF1 has two sub-units namely HIF-1 alpha (inducible) and HIF-1 beta (constitutive) which are basic helix-loop-helix proteins of the PAS family (Per, ARNT, Sim). HIF-1 alpha expression is regulated through alterations in cellular oxygen level as well as in an oxygen-independent manner via different cytokines (via PI3K-AKT-mTOR signaling), growth factors, oncogenic activation, or loss of tumor suppressor function. In normoxic cells, HIF-1 alpha gets proline hydroxylated which results in a conformational change promoting its binding to VHL protein E3 ligase complex, followed by its ubiquitination and rapid proteasomal degradation. Hypoxia as well as chemical hydroxylase inhibitors (desferrioxamine, cobalt, etc.) inhibit HIF-1 alpha degradation and lead to its accumulation in the cells which precedes its nuclear translocation and HIF1 target gene expression. Owing to its role in cellular signaling and hypoxia responsive transcription, HIF-1 alpha has been regarded as the 'Master Regulator of Hypoxia'.
What is HIF-2 alpha and how is it different from HIF-1 alpha?
HIF-2 alpha, also known as endothelial PAS domain protein-1 /EPAS1 or member of PAS superfamily 2/Mop2, is one of the three known heterodimeric hypoxia-inducible factors/HIFs family members. HIFs differ in their alpha subunits, but share a common beta subunit. Like HIF-1 alpha, the expression of HIF-2 alpha is regulated through hypoxia-dependent protein stabilization with the help of PHD proteins that initiate HIF2 degradation via the pVHL. The activity of PHD proteins is inhibited under hypoxic conditions leading to the accumulation of HIF1/HIF-2 alpha followed by nuclear translocation for the transcriptional activation. HIF-2 alpha shares ~48% amino-acid sequence homology with HIF-1 alpha and is now known to exert similar as well as divergent roles, especially in certain tumors. In vitro studies showed that HIF-1 alpha is mostly active 2-24 hours after hypoxic/anoxic treatments (<0.1% O2), while HIF-2 alpha continues to be active even after 2-3 days of physiological hypoxia (<5% O2) suggesting that HIF-1 participates in the initial response to hypoxia, while HIF-2 drives the hypoxic response during chronic hypoxic exposure (Koh and Powis 2012). HIF-2 alpha antibodies, which do not react with its homologue HIF-1 alpha, are essential to immunoassay-based analysis of HIF-2 alpha's role in hypoxia signaling. Bio-Techne offers high quality antibodies to various HIFs including HIF-1 Alpha, HIF-2 Alpha and HIF-3 Alpha.
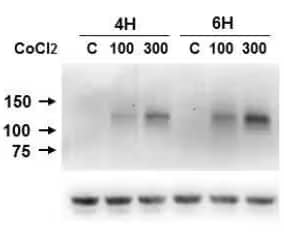
HIF-1 alpha Antibody (H1alpha67) [NB100-105]: Western blot (WB) analysis of lysates from Caki-1 cells which were left untreated or were treated with 100-300 μM of CoCl2 for 4-6 hours. The antibody detected single specific band of HIF-1 alpha at ~120 kDa. The blot was stripped and re-probed with beta-Actin as loading control (bottom image).
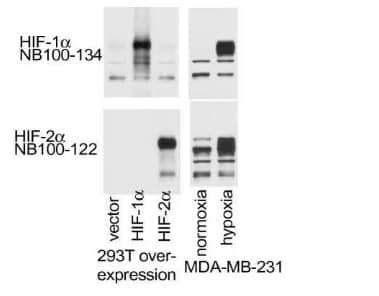
HIF-2 alpha/EPAS1 Antibody [NB100-122]: WB analysis of lysates from MDA-MB-231 cells which were subjected to normoxic/hypoxic conditions or transfection using empty, HIF1A or HIF2A overexpression vectors. This HIF-2 alpha antibody did not cross-react with HIF-1 alpha protein.
What is the role of HIF-3 alpha?
HIF-3 alpha (hypoxia-inducible factor 3-alpha) is the least studied member of HIF family. However, it is known to be regulated by HIF1 at the transcriptional level, and it exerts inhibitory effects on HIF-1 alpha or HIF-dependent gene regulation, in a cell-type specific manner. HIF‑3 alpha acts as a transcriptional regulator, which negatively affects the gene expression by competing with HIF-1 alpha and HIF-2 alpha in binding to target genes' transcriptional elements during hypoxia. Its role as transcription factor has been confirmed recently with the identification of HIF-3 alpha target genes (Yang et al 2015). Several alternative HIF-3 alpha splicing variants have been reported but their precise biological function has not been well defined. Bio-Techne's human and mouse targeted HIF-3 Alpha antibodies are highly specific and can be used to delineate the role of this protein in hypoxia dependent and independent signaling pathways.
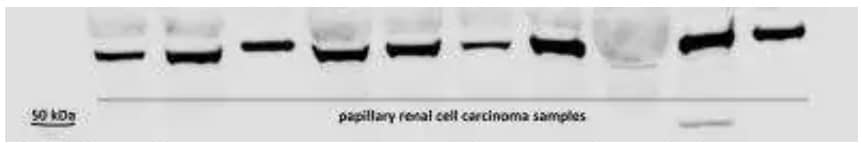
HIF-3 alpha Antibody [NB100-2529]: WB analysis of 30 μg lysates from human papillary renal cell carcinoma samples using HIF-3 alpha antibody at 1:1000 dilution with overnight incubation at 4˚C. Detection was performed using IRDye 800CW labeled secondary antibody on Odyssey Imaging System.
What is the significance of HIF-1 beta?
HIF-1 beta (also called as ARNT/aryl hydrocarbon receptor nuclear translocator) is a transcriptional subunit which forms heterodimer with AHR, AHRR, HIF-1 alpha and EPAS1/HIF-2 alpha as well as with other bHLH proteins. It has been shown to interact with TACC3 and NOCA7 also. HIF-1 beta dimerization is required for efficient transcription factors' binding to DNA. HIF-1 beta heterodimers with HIF-1 alpha or EPAS1/HIF-2 alpha act as transcriptional regulators of the adaptive response to hypoxia. Chromosomal translocations resulting in TEL-ARNT fusion protein have been shown to be associated with AML, and decreased HIF-1 beta expression levels in pancreatic islets from NIDDM subjects reflect its role in beta-cell function.
What size bands should I expect for HIF-1 alpha in Western blot assay?
The theoretical molecular weight of HIF-1 alpha is ~93kDa. However, post translationally modified HIF-1 alpha may run at 110-130 kDa. More specifically, the ubiquitinated HIF-1 alpha, which fails to undergo proteasomal degradation, will run between 110-130 kDa molecular weight range on a Western blot. Degraded HIF-1 alpha protein may show up at 40-60 kDa while the dimeric protein may appear at a position well above 200 kDa on non-reducing gels.
I do not detect HIF-1 alpha specific bands in lysates from normal or HIF1 overexpressing cells? Is it an antibody related problem?
Yes, could be an issue related to primary antibody, but it is helpful to consider a few points before drawing a conclusion. HIF-1 alpha is among the rapidly degradable proteins and it can degrade within the first 5 minutes of exposure to oxygen (Huang et al., 1996). This makes HIF-1 alpha a difficult target to study in Western blot assay. Studies have shown that HIF-1 alpha protein is barely detectable in normoxic cells, even when HIF-1 alpha is transiently overexpressed using pHIF-1a (expressing full-length HIF-1a mRNA), while an accumulation of HIF-1 alpha is observed under hypoxic conditions. This is because even the overexpressed HIF-1 alpha gets degraded under normoxic conditions. Besides weakening the specific signal, degradation leads to the detection of additional bands (at lower molecular weight). It is helpful to prepare lysates of cells as quickly as possible after the hypoxia exposure in a hypoxic incubator by scrapping the cells directly in lysis buffer. Moreover, it is generally advisable to analyze HIF-1 alpha in cytoplasmic and nuclear extracts instead of the whole cell lysates.
Why should one test HIF-1 alpha in nuclear-cytoplasmic fractions? Do you recommend any kit for nuclear-cytoplasmic fractions?
HIF-1 alpha localizes to the cytoplasm of the cells under normoxic conditions. However, under hypoxic conditions, it translocate to the nucleus to activate its target genes. Therefore, to analyze the active (nuclear) pool of HIF-1 alpha by Western blot, it is recommended to prepare nuclear-cytoplasmic fractions. Fractionation increases the amount of HIF-1 alpha in the mentioned lysates. Bio-Techne offers Nuclear Extraction Kit [NBP2-29447] which is a convenient tool to prepare nuclear-cytoplasmic fractions from mammalian cells and tissue samples.
What loading controls do you suggest for Nuclear/Cyto fractions?
Lamin B1, Histone H3 and Histone H1 are the most commonly used loading control markers for nuclear fractions. Beta Actin, alpha Tubulin, beta Tubulin, Glucose 6 Phosphate Dehydrogenase (G6PD) and GAPDH are good markers for use as loading controls for cytosolic fraction. It is pertinent to mention here that GAPDH is one of the target genes of HIF-1 alpha and if a change in its expression is observed, an alternative cytosolic control should be considered.
What is the significance of CoCl2-based controls?
CoCl2-treated cells are often used as controls in hypoxia signaling experiments. CoCl2 inhibits PHD enzymes (the oxygen sensors) through replacement of Fe with Co making these enzymes unable to mark HIF alpha for degradation (DFO shows similar effects on iron pool while DMOG also inhibits PHDs). It should be noted that CoCl2 only mimics HIF1 accumulation; hypoxia has other effects that are not mimicked by CoCl2 treatment. In addition to its effects on HIF-1 alpha accumulation, CoCl2 treatment can also modulate other enzymes and pathways (Triantafyllou et al., 2006). Bio-Techne offers ready to use true hypoxic/normoxic and hypoxia mimetic (CoCl2 induced) control lysates which are provided in Laemmli sample buffer with added beta-mercaptoethanol.
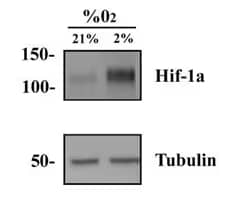
HeLa Hypoxic/Normoxic Lysate [NBP2-36452]
WB analysis of HeLa hypoxic and normoxic lysates using HIF-1 Alpha antibody (NB100-134). Hypoxic lysate showed several folds higher signal vs normoxic samples.
| Lysate | Catalog Number | Preparation Notes |
|---|---|---|
| HeLa (hypoxic/normoxic pair) | NBP2-36452 | Whole cell lysate of HeLa cells grown under true hypoxic conditions wherein the oxygen level in the incubator was reduced to 2% for 4 hours |
| HeLa (CoCl2 treated/untreated pair) | NBP2-36450 | Whole cell lysate of HeLa cells that were cultured using standard conditions and then exposed or not to 100 μM CoCl2 for 4 hours |
| HepG2 (hypoxic/normoxic pair) | NBP2-36453 | Whole cell lysate of HepG2 cells grown under true hypoxic conditions wherein the oxygen level in the incubator was reduced to 2% for 4 hours |
| HepG2 (CoCl2 treated/untreated pair) | NBP2-36451 | Whole cell lysate of HepG2 cells that were cultured using standard conditions and then exposed or not to 100 μM CoCl2 for 4 hours |
| Cos7 (CoCl2 treated/untreated pair) | NB800-PC26 | Nuclear lysate of COS-7 cells that were exposed or not to 16-hour incubation with 150 μM of CoCl2 |
What are the methods for stabilizing HIFs in cells?
CoCl2, DMOG (Catalog # 4408), DFO and MG 132 (Catalog # 1748) are some of the most commonly used HIF stabilizers for in vitro use. CoCl2 stabilizes HIF in its non-ubiquitinated form, whereas proteasome inhibitors such as MG 132 stabilize HIF in its ubiquitinated form. Since VLH regulates the degradation of HIFs, knockdown or knockout of VHL from cells can also be used for HIFs stabilization. However, hypoxic treatment (<5% O2) with a hypoxia chamber is the most physiological way to stabilize HIFs in cultured cells as well as in in vivo studies.
Do you have a standard or general protocol for generating hypoxic lysates?
Various cell lines may show a differential response to hypoxia. Therefore, users will need to optimize a threshold hypoxic treatment from cell line to cell line. Here are some general guidelines which may be considered as a starting point:
- Bring tissue culture incubator or chamber to 2% O2 or less, by the regulated addition of Nitrogen. If you are using a hypoxia chamber, you will need to consult the operation instructions for the addition of Nitrogen.
- Grow cells of choice to semi-confluency (70-80%) in 90 or150 mm culture dishes. Incubate the cells in the hypoxic incubator or chamber for several time points ranging from 2 -8 hours.
- Thereafter, aspirate the media from each dish, wash them with cold PBS on ice and lyse cells following the cell lysis protocol. Importantly, HIF1 alpha degrades within a couple of minutes under normal oxygen conditions, so a quicker processing of samples is recommended once the cells are out of hypoxic conditions.
How should I treat the cells for making CoCl2 based hypoxia mimetic lysates?
Here is a general protocol that we use in our lab for CoCl2:
- Grow cells in 90 or 150 mm tissue culture dishes to semi-confluency (70-80%).
- Prepare a fresh 100 mM stock solution of CoCl2 in PBS (e.g. 1.29mg of CoCl2 in PBS to make a total volume of 10mL). Add the CoCl2 solution to a final concentration of 100-150 μM directly to the tissue culture media in each dish (e.g. 10 μL of 100mM CoCl2 to 9.99mL of media will give a final concentration of 100 μM CoCl2) and mix well. Ensure to use a freshly prepared 100 mM CoCl2.
- Incubate the cells under standard tissue culture conditions (5% CO2, 37°C) for 4-8 hours.
- Aspirate the media from each dish and wash with PBS, aspirate all buffer as quickly as possible. Lyse cells following the cell lysis protocol.
Besides HIF-1 Alpha stabilizers, what are some other controls for hypoxia studies?
One can measure the protein or mRNA transcript expression levels of some hypoxia responsive genes such as CAIX, NF-kB, Glut1, PDK1 and VEGFA. Carbon Anhydrase IX /CAIX has been extensively used as a reliable marker for hypoxia induction, especially in experiments involving tissues analysis. Although HIFs are also great markers, the target genes/proteins suggested above tend to have longer half-lives which makes them more reliable for use as secondary/additional markers.
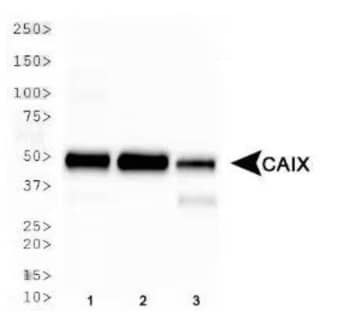
Carbonic Anhydrase IX Antibody [NB100-417] WB analysis of Carbonic Anhydrase IX expression in whole cell lysates of (1) HeLa, (2) MDA-MB-231 and (3) A549 cell lines.
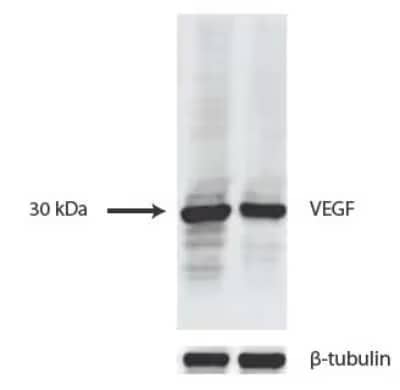
VEGF Antibody (VG1) [NB100-664] WB analysis of lysates from HUVEC (left) and 293T cells (right) using VEGF antibody at 1:1000 dilution.
In my samples, GAPDH expression also seems to be changing. What other options can I consider for loading control in Western blot?
Hypoxia has been shown to regulate GAPDH expression, so this is a very common observation. In this situation, it is advisable to consider a different loading control for normalizing the HIFs expression. Housekeeping genes such as beta-2 microglobulin (B2M) and ribosomal protein L13a (RPL13A) have been shown to be unchanged by hypoxia (Tang et al., 2010) and are suitable controls for HIF analysis.
In my repeated WB on the same samples, HIFs' bands in second run showed up at much higher position (top of the gel). Does it mean that the sample buffer caused aggregation of HIFs over the course of time?
Yes, changes in the ionic concentration due to freezing of the samples in sample buffer as well as the SDS mediated protein denaturation (which exposes the hidden hydrophobic groups) can contribute to protein aggregation. Therefore, the sample buffer is not an ideal buffer to store proteins for longer periods. To limit protein aggregation, we suggest preserving the samples in lysis buffer at -20 to -80˚C.
Are there any cell lines which have mutated VHL and/or over-expresses HIF-2 alpha?
Yes, the human 786-O cell line is a clear-cell renal carcinoma cell line /RCC cell line which constitutively expresses only HIF-2 alpha but not HIF-1 alpha or VHL. This cell line provides a great model for investigating the effects of both HIF isoforms, particularly HIF-2, on tumor growth and metabolism in in vitro as well as in vivo studies.
Does HIF expression vary from cell line to cell line? Also, can serum or glucose starvation have an impact on HIFs expression?
Yes, different cell lines do express different amounts of HIFs. The expression levels of HIFs are generally linked to the proliferative and/or invasive potential of the cell lines. Shi et al. 2010 have shown differential expression of HIF-1 alpha in human breast cancer cell lines, namely MDA-MB-231, MCF-7, SK-BR-3, MDA-MB-468 and MDA-MB-453 which were grown under serum starved conditions for 24 hours. With the help of 10-60 hour time points, they further showed enhancing effects of serum starvation on HIF-1 alpha expression in MDA-MB-231 cells which are highly aggressive in nature.
In cultured cells, can cell density affect the cellular response to hypoxia?
Yes, the cell density/confluency may have a direct impact on the extent of hypoxia induction in cell cultures. Confluent cells have higher oxygen consumption rate and hence, they are more prone to HIFs induction compared to low confluent cells.
What localization pattern can I expect for HIF-1 alpha in IHC or ICC assays?
HIF-1 alpha can be found at very low levels in the cytoplasm under normoxic conditions. During hypoxia, HIF-1 alpha is stabilized and translocates to the nucleus to act as a transcription factor. You should expect to see nuclear staining in your hypoxic samples, but may observe faint cytoplasmic staining in relation to degraded HIF in your samples.
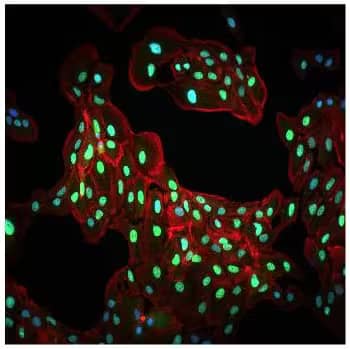
HIF-1 alpha Antibody [NB100-134]:ICC/IF analysis of HIF-1 alpha expression in ARPE-19 cells which were cultured under hypoxic conditions. The cells showed a mild cytoplasmic with very strong staining for HIF-1 alpha (green).
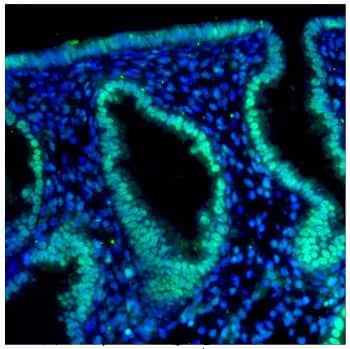
HIF-2 alpha Antibody (ep190b) [NB100-132]: IHC-P analysis of a formalin fixed tissue section of human endometrium using HIF-2 alpha/EPAS1 antibody (clone ep190b) at 1:250 dilution. The IHC assay involved an antigen retrieval step and blocking using 10% normal serum.
Is it necessary to perform antigen retrieval for HIF-1 alpha's IHC staining?
The need for an antigen retrieval step is mainly dependent on the nature of the fixative and the fixation time. When the tissues are fixed in formalin for more than a few hours, then antigen retrieval is required. Formalin cross-links the proteins which results in hindering of the antigenic sequences. Typically, paraffin or frozen sections from tissues fixed for 4 or more hours in formalin require an antigen retrieval step before labelling with primary antibodies. Antigen retrieval with 10 mM sodium citrate buffer (pH 6.0) is standard for many proteins (including HIFs). For more details on a given sample type, we recommend reviewing product citations for the antibodies under consideration.
For IHC-P analysis, what tissue should I use as a negative control for HIF-1 alpha?
HIF-1 alpha is highly expressed in most cancers as well as in normal kidney and heart tissues. Therefore, a normal tissue other than kidney and heart can be used as a negative control when performing IHC with HIF-1 alpha. To confirm the specificity of the observed staining, we also suggest the following controls in IHC-P assay: Negative control (no primary, no secondary), Secondary only negative control (no primary added), and Peptide block control (View our Peptide Competition Protocol).
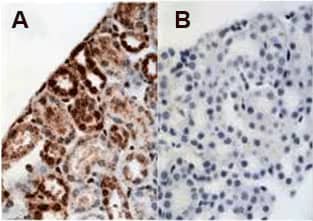
Recombinant Human HIF-1 alpha Protein [NBC1-18422]
IHC-P analysis of mouse kidney showing (A) staining with HIF-1 Alpha antibody NB100-479 and (B) NBC1-18422 as blocking control for NB100-479 (Images provided by Dr Yves Heremans, Vrije Universiteit Brussel, Brussels, Belgium).
For HIF-1 alpha Flow Cytometry, do I need to permeabilize the cells?
HIF-1 alpha mainly localizes to the cytoplasm, but under hypoxic conditions, it translocates to the nucleus. To analyze cytoplasmic and nuclear pools of HIF-1 alpha, permeablization is required. Permeablization for flow cytometry can be achieved by incubating the cells with cold 80% methanol (after 2% paraformaldehyde fixation) as explained in by Pantel et al. in PLoS Biol. 2014.
Do you offer any inhibitors or inducers of HIF-1/HIF-2 α or hypoxia signaling?
Bio-Techne offers a wide range of small molecule inhibitors and inducers of hypoxia signaling proteins:
| Catalog # | Name | Activity |
|---|---|---|
| 4705 | Chetomin | Inhibits the interaction of HIF-1/-2α and STAT2 with CBP/p300 by targeting CBP/p300’s CH1 domain. |
| 5582 | TAT-cyclo-CLLFVY | Inhibits HIF-1 (but not HIF-2) α dimerization with HIF-1β. |
| 4408 | DMOG | Increase HIF-1α by inhibiting HIF-α Prolyl 4-hydroxylase/ P4H enzyme. |
| 4324 | KC7F2 | Inhibits HIF-1α through its translational down-regulation |
| 5243 | TC-S 7009 | HIF-2α/EPAS1 inhibitor with a high affinity and specificity |
Find more hypoxia-related small molecules.
Do you have info about using HIF-1 alpha in Western Blot?
- The HIF proteins are among the most rapidly degrading proteins ever studied. Upon cellular re-oxygenation it can be completely degraded in less than 1 minute. Therefore, it is critical to prep only a few plates/dishes/flasks of cells at a time and to immediately place the cells into ice cold buffers and perform the whole protein prep on ice.
- HIF-1 is largely undetectable in cells or tissues grown under normoxic conditions. It is stabilized only at O2 concentrations below 5% or with treatment using certain agents (CoCl2, DFO, etc.) so proper sample preparation is critical.
- Upon stabilization HIF-1 translocates to the nucleus. The best western blots (cleanest) are always done using nuclear extracts. It is possible to detect HIF-1 in whole cell extracts, but they tend to be much dirtier and the staining is much weaker.
- We recommend that a positive/negative control always be run side by side so that it is possible to discern which band is upregulated in the hypoxic sample. Unprocessed HIF1 is ~95 kDa while the fully post-translationally modified form is ~116 kDa, or larger. Additionally, HIF-1 alpha may form a heterodimer with HIF-1 beta (Duan, et al. (2005) Circulation 111: 2227-2232)8. Depending on the sample, treatment, etc. you may see either a band or a doublet.
- Bio-Techne offers hypoxic and normoxic cell lysates for positive controls in your western blot experiments. Both CoCl2-induced and true hypoxia-induced cell lysates are available.
- Huang LE, Arany Z, Livingston DM, Bunn HF. (1996) Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996 Dec 13;271(50):32253-9.
- Pantel A, Teixeira A, Haddad E et al. (2014) Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol. 12(1):e1001759.
- Shi Y, Chang M, Wang F et al. (2010) Role and mechanism of hypoxia-inducible factor-1 in cell growth and apoptosis of breast cancer cell line MDA-MB-231. Oncol Lett. 2010 Jul;1(4):657-662
- Koh MY, & Powis G. (2012) Passing the baton: the HIF switch. Trends Biochem Sci. 37(9):364-72.
- Yang SL, Wu C, Xiong ZF, & Fang X. (2015) Progress on hypoxia-inducible factor-3: Its structure, gene regulation and biological function (Review). Mol Med Rep. 12(2):2411-6.
- Triantafyllou A, Liakos P, Tsakalof A et al. (2006) Cobalt induces hypoxia-inducible factor-1alpha (HIF-1alpha) in HeLa cells by an iron-independent, but ROS-, PI-3K- and MAPK-dependent mechanism. Free Radic Res. 40(8):847-56.
- Tang K, Xia FC, Wagner PD, & Breen EC. (2010). Exercise-induced VEGF transcriptional activation in brain, lung and skeletal muscle. Respir Physiol Neurobiol. 170(1):16-22
- Duan, et al. (2005) Endothelium-Intrinsic Requirement for Hif-2α During Vascular Development. Circulation 111:2227-2232.