Bacterial Pathogens
Bacterial Classification
Bacteria are classified within two major groups, Gram-positive or Gram-negative, based on their cell wall composition. Peptidoglycan is a main component of the cell wall that surrounds the cell membrane, provides protection from osmotic changes, and determines the bacterial cell shape. The rigid mesh-like peptidoglycan layer consists of two types of glycans, N-acetylglucosamine and N-acetylmuramic acid, connected by short peptides. In Gram-positive bacteria the peptidoglycan mesh is wider, consisting of multiple layers and associated teichoic acids, polysaccharides, and peptidoglycolipids. In contrast, Gram-negative bacteria have a thin cell wall with less peptidoglycan content and cross-linking. Gram-negative bacteria are additionally surrounded by an outer membrane containing lipopolysacharides (LPS) and lipoproteins.
Gram-Positive and Gram-Negative Bacteria
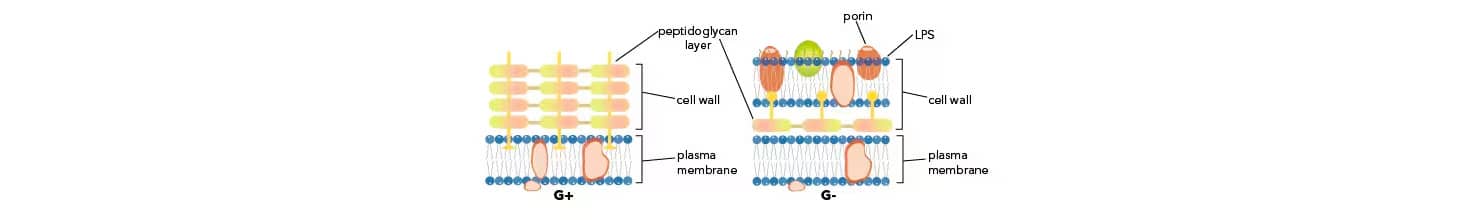
Bacterial envelope composition distinguishes two major bacterial groups. The Gram staining technique was developed by the Danish bacteriologist Hans Christian Gram. This technique identifies bacteria based on their cell wall composition and allows classification of most bacteria as Gram-positive or Gram-negative. Briefly, during Gram staining bacteria are exposed to crystal violet dye and iodine which results in the formation of a complex, crystal violet-iodine complex. A solvent step following dye treatment, dehydrates the thicker peptidoglycan layer trapping the dye and staining bacteria, thereby referred to as Gram-positive. In contrast, solvent treatment of bacteria with a thin peptidoglycan layer, dissolves the outer membrane and allows the dye to leach out, thereby referred to as Gram-negative.
Gram-Positive Bacterial Pathogens
Gram-Negative Bacterial Pathogens
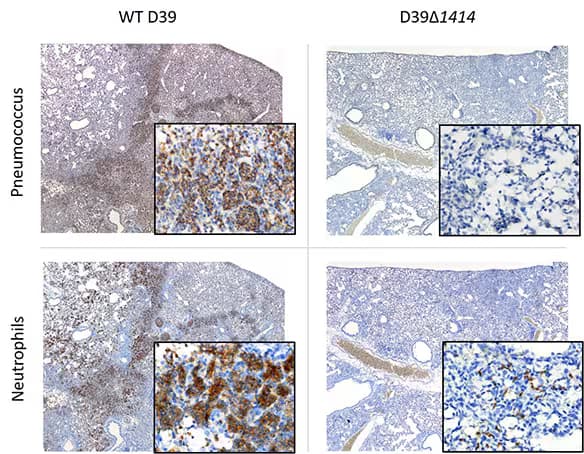
The pathogen, Streptococcus pneumoniae (pneumococcus), increases morbidity and mortality of influenza-infected hosts. Histopathology of lung sections collected at 24 h pbi from mice co-infected with mouse adapted influenza A/Puerto Rico/8/34 (H1N1) (PR8) and type 2 pneumococcal strain D39 variants. Serial lung sections were subjected to immunohistochemistry (IHC) for pneumococcus using Rabbit Anti-Streptococcus pneumoniae Polyclonal Antibody (Catalog # NB100-64502) at 4 µg/mL or neutrophils using Rat Anti-Ly-6G6C Monoclonal Antibody (Catalog # NB600-1387) at 2 µg/mL. Representative images at 4x magnification with 60x magnification inset are shown. Images courtesy of Dr. Amanda P. Smith, UTHSC, TN //doi.org/10.1101/659557
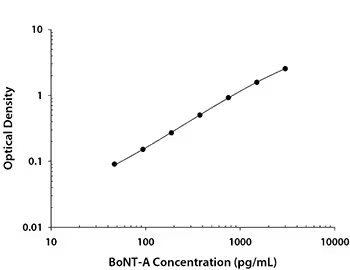
Solid phase sandwich ELISA, Botulinum Neurotoxin Type A DuoSet ELISA, 5 plate (DY4489-05). Botulinum neurotoxin is produced by Clostridium botulinum and consists of a light A- and heavy protein B-subunit. The A-subunit inhibits the release of acetylcholine at neuromuscular junctions.
Mycoplasma and Mycobacterium: How are they different?
Mycoplasma species are Gram-negative bacteria which lack a cell wall. The Mycoplasma genus contains over a 100 species, some of which lead to human disease including Mycoplasma pneumoniae and Mycoplasma genitalium. In contrast, Mycobaterium species are characterized by a complex cell wall, consisting of a mycolyl-arabinoglactan-peptidoglycan complex, which sets them apart from other Gram positive bacteria. The presence of an outer membrane “myco-membrane” is a feature that Mycobacterium species share with Gram negative bacteria. Close to 200 Mycobacterium species have been identified including important human pathogens such as Mycobacterium tuberculosis and Mycobacterium leprae.
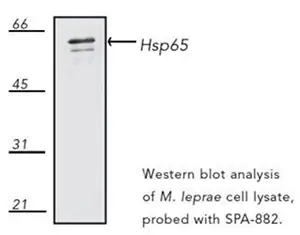
The 65-kDa heat shock protein (HSP65) is a predominant component of Mycobacterium leprae’s cell wall. HSP65 is highly immunogenic and recognized by T cells. Western blot analysis of the HSP65 expression in Mycobacterium leprae cell lysate using a mouse monoclonal antibody raised against Mycobacterium bovis BCG Hsp65, Hsp65 (mycobacterial) Antibody (4H11) [NBP1-97874].
Fluorescent Probes to Image Gram-Positive and Gram-Negative Bacteria
Incorporation of fluorescently-labeled metabolic precursors by live bacteria results in the labeling of specific cell wall and envelope components such as peptidoglycans and trehalose glycolipids, respectively. In metabolic labeling, fluorescent reporters target specific cell surface structures allowing investigators to study bacterial growth and responses to pharmacological agents. Combined with super-resolution microscopy, fluorescent metabolic labeling enables the analysis of cell wall and envelope dynamics.
| Fluorescent Probe | Description | Excitation/Emission Wavelength (nm) |
|---|---|---|
| HADA | Blue fluorescent D-amino acid, labels peptidoglycan in live bacteria. | 405/460 |
| NADA-green | Green fluorescent D-amino acid, labels peptidoglycan in live bacteria. | 450/555 |
| RADA | Orange-red fluorescent D-amino acid, labels peptidoglycan in live bacteria. | 554/580 |
| sBADA | Green sulfonated BODIPY-FL 3-amino-D-alanine (sBADA), labels peptidoglycan in live bacteria. | 490/510 |
| 6 TMR Tre | Fluorescent trehalose, selectively labels mycobacterial cell envelope. | 532/580 |
| YADA | Green-yellow lucifer yellow-based fluorescent D-amino acid, labels peptidoglycan in live bacteria. | 426/535 |
Find all Fluorescent Probes for Imaging Bacteria.
Antibiotics
Treatment of bacterial infections relies primarily on the use of antibiotics, which are chemicals that inhibit bacterial growth or kill bacteria. Based on their main mechanism of action, antibiotics are classified as bacteriostatic or bactericidal, depending on whether they stop bacterial growth or induce cell death, respectively. Beyond these major mechanisms, bactericidal antibiotics may induce cell death by targeting and inhibiting the synthesis of DNA, RNA, protein or cell wall components.
| Antibiotic | Description | Mechanism |
|---|---|---|
| Doxycycline hyclate | Broad-spectrum antibiotic. Tetracycline derivative. Inhibits bacterial protein synthesis. Binds bacterial 30S ribosomal subunit. | Bacteriostatic |
| Gentamicin sulfate | Aminoglycoside antibiotic. Irreversibly binds bacterial ribosomes and disrupts protein synthesis. Active against gram-positive and -negative bacteria. Stable over wide range of temperatures. | Bactericidal |
| Tobramycin | Aminoglycoside antibiotic. Exhibits antibacterial activity against strains of Enterobacteriaceae, Pseudomonas and Staphylococcus aureus. | Bactericidal |
| Trovafloxacin mesylate | Fluoroquinolone antibiotic. Inhibits bacterial DNA synthesis through inhibition of bacterial DNA topoisomerase IV and DNA gyrase. | Bacteriostatic |
Browse Antibiotics available from Bio-Techne.
Antibiotic Resistance
Development of antibiotic resistant bacteria is currently a major public health concern. Antibiotic resistance mechanisms are diverse and encoded by bacterial genomic programs and extra-chromosomal or plasmid DNA. Several in vitro methods allow assessment of bacterial resistance to antibiotics including:
Disc diffusion testing- Involves culture of bacterial specimens on agar plates, followed by exposure to antibiotics through placement of discs containing antibiotic drug. Inhibition of growth around the antibiotic containing disc is used as a measure of antibiotic susceptibility.
Minimum inhibitory concentration- Involves inoculation of bacteria in serial dilutions of an antibiotic, typically broth, to determine the minimal antibiotic concentration which inhibits visible bacterial growth. This is based on the evaluation of bacterial growth following overnight culture.
Mechanisms of Antibiotic Resistance
Bacterial antibiotic resistance may be acquired by spontaneous genetic mutations or transfer of genetic material from other bacteria or bacteriophages. Various mechanisms of antibiotic resistance have been identified and bacteria may exhibit a single or multiple antibiotic resistance traits.
| Antibiotic: Resistance Mechanisms | Bacterial Proteins |
|---|---|
| Enzyme mediated chemical modification reduces or abolishes antibiotic activity | Hydrolases (e.g., NDM-1); Transferases (e.g., Neomycin phosphotransferase II); Redox enzymes (e.g., Tetracycline monohydroxylase TetX) |
| Reduction of antibiotic uptake | Porins (aqueous channels, provide route for antibiotics into periplasmic space), mutations lead to reduced drug uptake (e.g., E. coli's OmpC, OmpF mutations and β-lactam resistance; Helicobacter pylori OMPs alterations and clarithromycin resistance) |
| Increased antibiotic efflux | Multidrug efflux pumps (e.g., Helicobacter pylori’s RND efflux pumps- HefABC, HefDEF, and HGHI; Meticillin-resistant Staphylococcus aureus' s (MRSA) NorB efflux pump overexpression) are membrane proteins involved in extrusion of multiple types of antibiotics |
| Increased antibiotic target | Overexpression of antibiotic target leading to increased minimal inhibitory antibiotic concentration (e.g., E. coli dihydrofolate reductase overexpression leads to trimethoprim resistance). |
| Change in bacterial antibiotic target (e.g., mutations, recombination events) | Reduced or blocked antibiotic binding to key functional targets (e.g., Enterococcus vancomycin resistance due to decreased antibiotic binding to peptidoglycan precursors) |
Major Bacterial Human Pathogens
In 2017 WHO published a list of "priority pathogens" which includes twelve bacterial families with increased pathogenic potential to humans due to their increased incidence of antibiotic resistance. The list classifies pathogens as critical, high and medium based on the global need for the development of new antibiotics.
| *Priority Pathogens | Antibiotic Resistance | Description/Groups Affected |
|---|---|---|
| Critical Priority | ||
| Acinetobacter baumannii | Carbapenem-resistant | Opportunistic pathogen found in water and soil, affects immunocompromised patients leading to infection (e.g., blood, urinary, and pneumonia). |
| Pseudomonas aeruginosa | Carbapenem-resistant | Opportunistic pathogen found in water and soil, common in post-surgery associated infections (e.g., blood, and pneumonia). |
| Enterobacteriaceae |
Carbapenem-resistant Extended spectrum β-lactamase producing |
Common infectious agent in hospital settings (e.g., E. coli, Klebsiella pneumoniae). |
| High Priority | ||
| Enterococcus faecium | vancomycin-resistant | Common infectious agent in hospital settings, frequently affects elderly and immunocompromised patients (e.g., urinary and blood infections). |
| Staphylococcus aureus | methicillin-resistant, vancomycin-intermediate and resistant | Community or health care associated infectious agent, spreads through contact with infected people or objects (e.g., skin infection through open wounds). |
| Helicobacter pylori | clarithromycin-resistant | Frequently infects the stomach in an estimated 40% of people and may be asymptomatic or lead to peptic ulcers. |
| Campylobacter spp. | fluoroquinolone-resistant | Common foodborne pathogen, infections usually occur through ingestion of contaminated foods (e.g., undercooked poultry, cross-contaminated foods). |
| Salmonella | fluoroquinolone-resistant | Common foodborne pathogen, infections occur frequently in association with contaminated foods (e.g., raw or undercooked meats and eggs). |
| Medium Priority | ||
| Streptococcus pneumoniae | penicillin-non-susceptible | Upper respiratory tract commensal, may cause infection under conditions of host susceptibility (e.g., middle ear, lungs, blood, and meninges). |
| Haemophilus influenzae | ampicillin-resistant | Upper respiratory tract commensal, may cause infection under conditions of host susceptibility (e.g., middle ear, lungs, blood, and meninges). |
| Shigella spp. | fluoroquinolone-resistant | Foodborne pathogen, infection results through contaminated foods and as the result of poor hand washing practices leading to intestinal disease. |
*From WHO priority pathogens list for research and development of new antibiotics.
Helicobacter pylori binds to the gastric epithelium inducing inflammatory responses. Urase produced by H. pylori supports bacterial colonization and survival within the gastric mucosa. Urease also functions as a virulence factor and is implicated in the inflammatory process through various mechanisms (e.g., activation of CD74 in gastric epithelium and induction of IL8 production).
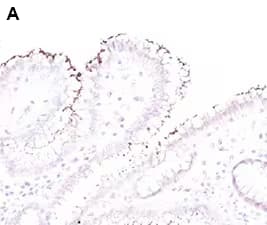
Immunohistochemical analysis of Helicobacter pylori in Formalin-fixed paraffin-embedded human stomach tissue sections with a rabbit polyclonal antibody to Helicobacter pylori lysate, Helicobacter pylori Antibody [NBP2-29479]
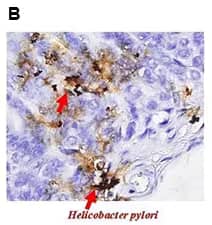
Immunohistochemical staining of Helicobacter pylori urase B in tissue from mice gastric pylori with a rabbit polyclonal antibody, Helicobacter pylori urease B. [NBP2-42850]
Anti-virulence Strategies to Target Multi-Drug Resistant Pathogens
Quorum sensing (QS) signaling is a mechanism of bacterial communication, which allows regulation of virulence factors, biofilm formation, colonization, and environmental adaptation. Bacterial communication is underscored by the release and extracellular accumulation of autoinducer molecules or AIs (e.g., autoinducer-2, acylated homoserine lactones (acyl-HSLs), oligopeptides, Pseudomonas quinolone signal molecule, diffusible signal factor, γ-butyrolactone, and 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde). Increase of extracellular AI concentration is dependent on bacterial density and leads to the activation of signaling pathways which regulate the expression of bacterial virulence genes. Targeting QS signaling has been proposed as a potential anti-bacterial strategy which may circumvent the development of bacterial resistance. Various strategies to inhibit QS depend on inhibition of AI expression or synthesis, inhibition of AI extracellular accumulation, and blockade of AI detection.
View our Quorum Sensing Signaling modulators.
Abushaheen, M. A., et al. (2020). Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month. https://doi.org/10.1016/j.disamonth.2020.100971
Alcalde-Rico, M., Hernando-Amado, S., Blanco, P., & Martïnez, J. L. (2016). Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2016.01483
Alderwick, L. J., Harrison, J., Lloyd, G. S., & Birch, H. L. (2015). The mycobacterial cell wall—peptidoglycan and arabinogalactan. Cold Spring Harbor Perspectives in Medicine. https://doi.org/10.1101/cshperspect.a021113
Balish, M. F. (2014). Mycoplasma pneumoniae, an underutilized model for bacterial cell biology. Journal of Bacteriology. https://doi.org/10.1128/JB.01865-14
Barker, K. F. (1999). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2014301/Antibiotic resistance: A current perspective. British Journal of Clinical Pharmacology. https://doi.org/10.1046/j.1365-2125.1999.00997.x
Beswick, E. J., Pinchuk, I. V., Minch, K., Suarez, G., Sierra, J. C., Yamaoka, Y., & Reyes, V. E. (2006). The Helicobacter pylori urease B subunit binds to CD74 on gastric epithelial cells and induces NF-κB activation and interleukin-8 production. Infection and Immunity. https://doi.org/10.1128/IAI.74.2.1148-1155.2006
Blanco, P., Hernando-Amado, S., Reales-Calderon, J., Corona, F., Lira, F., Alcalde-Rico, M., … Martinez, J. (2016). Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms. https://doi.org/10.3390/microorganisms4010014
Courvalin, P. (2006). Vancomycin Resistance in Gram-Positive Cocci. Clinical Infectious Diseases. https://doi.org/10.1086/491711
Egan, A. J. F., Cleverley, R. M., Peters, K., Lewis, R. J., & Vollmer, W. (2017). Regulation of bacterial cell wall growth. FEBS Journal. https://doi.org/10.1111/febs.13959
Egorov, A. M., Ulyashova, M. M., & Rubtsova, M. Y. (2018). Bacterial enzymes and antibiotic resistance. Acta Naturae. https://doi.org/10.32607/20758251-2018-10-4-33-48
Fernández, L., & Hancock, R. E. W. (2012). Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clinical Microbiology Reviews. https://doi.org/10.1128/CMR.00043-12
Fleitas Martínez, O., Rigueiras, P. O., Pires, Á. da S., Porto, W. F., Silva, O. N., de la Fuente-Nunez, C., & Franco, O. L. (2018). Interference With Quorum-Sensing Signal Biosynthesis as a Promising Therapeutic Strategy Against Multidrug-Resistant Pathogens. Frontiers in Cellular and Infection Microbiology. https://doi.org/10.3389/fcimb.2018.00444
Flensburg, J., & Skold, O. (1987). Massive overproduction of dihydrofolate reductase in bacteria as a response to the use of trimethoprim. European Journal of Biochemistry. https://doi.org/10.1111/j.1432-1033.1987.tb10664.x
Hentzer, M., Wu, H., Andersen, J. B., Riedel, K., Rasmussen, T. B., Bagge, N., … Givskov, M. (2003). Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO Journal. https://doi.org/10.1093/emboj/cdg366
Jiang, Q., Chen, J., Yang, C., Yin, Y., Yao, K., & Song, D. (2019). Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. BioMed Research International. https://doi.org/10.1155/2019/2015978
Kohanski, M. A., Dwyer, D. J., & Collins, J. J. (2010). How antibiotics kill bacteria: From targets to networks. Nature Reviews Microbiology. https://doi.org/10.1038/nrmicro2333
Kwak, Y. G., Truong-Bolduc, Q. C., Kim, H. Bin, Song, K. H., Kim, E. S., & Hooper, D. C. (2013). Association of norb overexpression and fluoroquinolone resistance in clinical isolates of Staphylococcus aureus from Korea. Journal of Antimicrobial Chemotherapy. https://doi.org/10.1093/jac/dkt286
McDermott, P. F., Walker, R. D., & White, D. G. (2003). Antimicrobials: Modes of action and mechanisms of resistance. International Journal of Toxicology. https://doi.org/10.1080/10915810305089
Palliyil, S., Downham, C., Broadbent, I., Charlton, K., & Porter, A. J. (2014). High-Sensitivity Monoclonal Antibodies Specific for Homoserine Lactones Protect Mice from Lethal Pseudomonas aeruginosa Infections. Applied and Environmental Microbiology. https://doi.org/10.1128/AEM.02912-13
Parada, C. A., Portaro, F., Marengo, E. B., Klitzke, C. F., Vicente, E. J., Faria, M., … Fernandes, B. L. (2011). Autolytic Mycobacterium leprae Hsp65 fragments may act as biological markers for autoimmune diseases. Microbial Pathogenesis. https://doi.org/10.1016/j.micpath.2011.06.001
Rémy, B., Mion, S., Plener, L., Elias, M., Chabrière, E., & Daudé, D. (2018). Interference in bacterial quorum sensing: A biopharmaceutical perspective. Frontiers in Pharmacology. https://doi.org/10.3389/fphar.2018.00203
Salton MRJ, Kim KS. Structure. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 2. Available from: https://www.ncbi.nlm.nih.gov/books/NBK8477/